
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,693,273 – 14,693,373 |
| Length | 100 |
| Max. P | 0.687652 |

| Location | 14,693,273 – 14,693,373 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.44645 |
| G+C content | 0.45223 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -16.53 |
| Energy contribution | -18.95 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14693273 100 - 21146708 GAUAGUGACUAGAUAGUUUUAGUGAU-----GUGCCCAUGAUGUCACGCCGCAGGUGAUAAAUCAGCGCUACUGUUGGAGUCGCCUGCAAAUGAUAACACAAUUG- ....(((..............(((((-----((.......)))))))...(((((((((...((((((....)))))).))))))))).........))).....- ( -28.30, z-score = -1.52, R) >droAna3.scaffold_13266 2222208 77 + 19884421 ---------GAUAUACUUUUAU---------GACCAUAUGAUGUCACUAAAGCUGCGAUAAAUCAUCGCUACUG--GCACUCCGCUGCAAAUGAUAA--------- ---------......((((..(---------(((........))))..))))..(((((.....))))).....--(((......))).........--------- ( -12.20, z-score = -0.52, R) >droEre2.scaffold_4845 8885365 94 - 22589142 GAUAGUGACUGGGUAGUUUUGGU---------AGCCCGUGAUGUCACGCCGCAGGUGAUAAAUCAGCGCAGCUGCUGGGGUCGCCUGCAGAUGAUAACAC--UUG- ..........((((.........---------.))))(((.(((((..(.(((((((((...((((((....)))))).))))))))).).))))).)))--...- ( -35.40, z-score = -1.73, R) >droYak2.chr2R 6636019 104 + 21139217 GAUAGUGACUAUGUAGUUUUAGUAGUUUUAGUAGUCCGUGAUGUCACGCCGCUGGUAAUAAAUCAGCGCUACUGUUAGGGUCGCCUGCAAAUGAUAACAC--UUGU ....((((((.........(((((((..........((((....))))..((((((.....)))))))))))))....))))))..((((.((....)).--)))) ( -25.62, z-score = -0.22, R) >droSec1.super_1 12201866 98 - 14215200 GAUAGUGACUAGAUAGUUUUAGUGA-------UGCCCAUGAUGUCACGCCGCAGGUGAUAAAUCAGCGCUAUUGUUGGAGUCGCCUGCAGAUGAUAACACAAUUG- ....(((((((((....))))))..-------.........(((((..(.(((((((((...((((((....)))))).))))))))).).))))).))).....- ( -28.70, z-score = -1.90, R) >droSim1.chr2R 13411735 98 - 19596830 GAUAGUGACUAGAUAGUUUUAGUGA-------UGCCCAUGAUGUCACGCCGCAGGUGAUAAAUCAGCGCUACUGUUGGAGUCGCCUGCAGAUGAUAACACAAUUG- ....(((((((((....))))))..-------.........(((((..(.(((((((((...((((((....)))))).))))))))).).))))).))).....- ( -28.70, z-score = -1.75, R) >consensus GAUAGUGACUAGAUAGUUUUAGUGA_______AGCCCAUGAUGUCACGCCGCAGGUGAUAAAUCAGCGCUACUGUUGGAGUCGCCUGCAAAUGAUAACAC__UUG_ ....(((((((((....))))))..................(((((....(((((((((...((((((....)))))).)))))))))...))))).)))...... (-16.53 = -18.95 + 2.42)
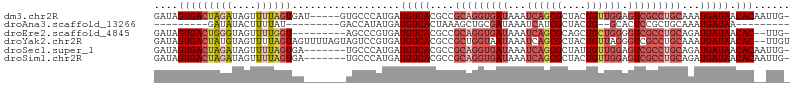
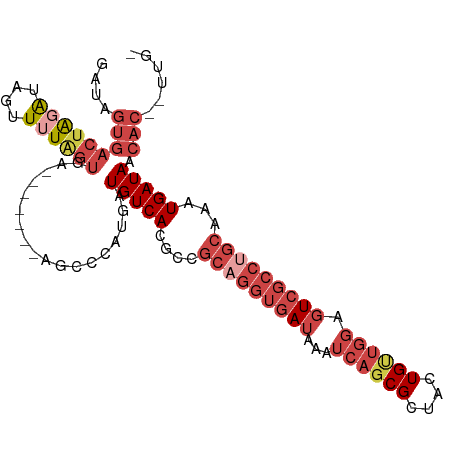

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:49 2011