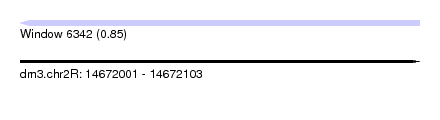
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,672,001 – 14,672,103 |
| Length | 102 |
| Max. P | 0.848669 |

| Location | 14,672,001 – 14,672,103 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.59781 |
| G+C content | 0.53522 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.61 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
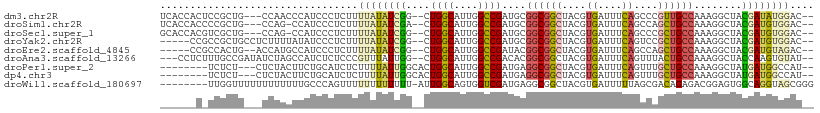
>dm3.chr2R 14672001 102 - 21146708 UCACCACUCCGCUG---CCAACCCAUCCCUCUUUUAUAUCGG--CUGGCAUUGGCCGAUGCGGCGGCUACGUGAUUUCAGCCCGUUGCCAAAGGCUACGAUAUGGAC-- ((((....((((((---(...................(((((--((......))))))))))))))....)))).((((...(((.(((...))).)))...)))).-- ( -32.40, z-score = -1.14, R) >droSim1.chr2R 13388769 101 - 19596830 UCACCACCCCGCUG---CCAG-CCAUCCCUCUUUUAUAUCGA--CUGGCAUUGGCCGAUGCGGCGGCUACGUGAUUUCAGCCAGCUGCCAAAGGCUACGAUGUGGAC-- ...((((..((.((---((((-.(................).--)))))).(((((...(((((((((..........)))).)))))....)))))))..))))..-- ( -33.99, z-score = -1.35, R) >droSec1.super_1 12180719 101 - 14215200 GCACCACGUCGCUG---CCAG-CCAUCCCUCUUUUAUAUCGG--CUGGCAUUGGCCGAUGCGGCGGCUACGUGAUUUCAGCCCGCUGCCAAAGGCUACGAUGUGGAC-- ...((((((((.((---((((-((................))--)))))).(((((...(((((((((..........)).)))))))....)))))))))))))..-- ( -47.39, z-score = -4.10, R) >droYak2.chr2R 6614243 100 + 21139217 -----CCGCCGCUGCCUCUUUUAUAUCCCUCUUUUAUAUCGG--CUGGCAUUGGCCGAUGCGGCGGCUACGUGAUUUCAGUCCGCUGCCAAAGGCUACGAUGUGGAC-- -----..((((((((......((((.........))))((((--((......)))))).))))))))............((((((.(((...)))......))))))-- ( -34.80, z-score = -1.52, R) >droEre2.scaffold_4845 8864256 98 - 22589142 -----CCGCCACUG--ACCAUGCCAUCCCUCUUUUAUAUCGG--CUGGCAUUGGCCGAUACGGCGGCUACGUGAUUUCAGCCAGCUGCCAAAGGCUACGAUGUAGAC-- -----.........--................((((((((((--(((((..((((((......))))))..((....)))))))).(((...)))..))))))))).-- ( -31.30, z-score = -1.19, R) >droAna3.scaffold_13266 2198332 102 + 19884421 ---CCUCUUUGCCGAUAUCUAGCCAUCUCUCCCGUUUAUUGG--CUGGCAUUGGCCGACACGGCGGCUACGUGAUUUCAGUUUACUGCCAAAGGCUACCAAGUGUAU-- ---.......((((((..(((((((..............)))--)))).))))))..(((((((((..((.((....))))...)))))...((...))..))))..-- ( -29.44, z-score = -1.07, R) >droPer1.super_2 5464156 96 + 9036312 --------UCUCU---CUCUACUUCUGCAUCUCUUUUAUUGGCACUGGCAUUGGCCGAUGAGGCGGCUACGUGAUUUCAGUUUGCUGCCAAAGGCUAUGAUGGCCAU-- --------.....---...................((((((((.(.......)))))))))((((((.((.((....))))..))))))...((((.....))))..-- ( -27.10, z-score = -0.48, R) >dp4.chr3 16400496 96 + 19779522 --------UCUCU---CUCUACUUCUGCAUCUCUUUUAUUGGCACUGGCAUUGGCCGAUGAGGCGGCUACGUGAUUUCAGUUUGCUGCCAAAGGCUAUGAUGGCCAU-- --------.....---...................((((((((.(.......)))))))))((((((.((.((....))))..))))))...((((.....))))..-- ( -27.10, z-score = -0.48, R) >droWil1.scaffold_180697 2162112 100 - 4168966 --------UUGGUUUUUUUUUUUUGCCCAGUUUUUUUUUUUU-AUUGGCAGUGGUCGAUGAGGCGGCUACGUGAUUUUUAGCGACAGAGACGGAGUGGCAGGUAGCGGG --------.................(((..........((((-((((((....))))))))))..(((((.((..((((..(......)..))))...)).)))))))) ( -22.50, z-score = -0.12, R) >consensus ______C_UCGCUG___CCUACCCAUCCCUCUUUUAUAUCGG__CUGGCAUUGGCCGAUGCGGCGGCUACGUGAUUUCAGCCCGCUGCCAAAGGCUACGAUGUGGAC__ .................................((((((((....((((....))))....((((((....((....))....))))))........)))))))).... (-15.49 = -15.61 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:48 2011