| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,643,833 – 14,643,924 |
| Length | 91 |
| Max. P | 0.756977 |

| Location | 14,643,833 – 14,643,924 |
|---|---|
| Length | 91 |
| Sequences | 14 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 57.81 |
| Shannon entropy | 0.92280 |
| G+C content | 0.40416 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -9.13 |
| Energy contribution | -8.73 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14643833 91 + 21146708 AAGAAGGUGGCUGUGACAAUGGUUAUGGACAUGCUUAUGGCUGGAAUUGAUACGGUAUGAUUAUUAAU---AUAUUUGGUACGAUUCAAUACUA--------------- .....(((((((((((..(((........)))..)))))))).((((((.(((((((((........)---)))))..)))))))))...))).--------------- ( -16.50, z-score = -0.45, R) >droSim1.chr2R 13360609 91 + 19596830 AAGAAGGUGGCUGUGACAAUGGUUAUGGACAUGCUUAUGGCUGGAAUCGAUACGGUAUGAUUAUUAAU---AUAUUCGGUACGAUUCAAUACUA--------------- .....(((((((((((..(((........)))..)))))))).((((((...(((((((........)---))).)))...))))))...))).--------------- ( -18.70, z-score = -0.68, R) >droSec1.super_1 12152663 91 + 14215200 AAGAAGGUGGCUGUGACAAUGGUUAUGGACAUGCUUAUGGCUGGAAUCGAUACGGUAUGAUUAUUGAU---AUAUUCGGUACGAUUUAAUACUA--------------- .....(((((((((((..(((........)))..)))))))).((((((...(((((((........)---))).)))...))))))...))).--------------- ( -16.50, z-score = -0.04, R) >droYak2.chr2R 6585393 91 - 21139217 AAGAAGGUGGCUGUGACAAUGGUUAUGGACAUGCUAAUGGCCGGAAUUGAUACGGUAUGAGAUAUGAC---AUAGACGGCAGGAUUUACAACUA--------------- ..(((....(((((........(((((..((((((....((((.........))))...)).)))).)---)))))))))....))).......--------------- ( -16.70, z-score = -0.30, R) >droEre2.scaffold_4845 8835613 91 + 22589142 AAGAAGGUGGCUGUGACCAUGGUUAUGGACAUGCUUAUGGCCGGAAUUGAUACGGUAAGAGAUACAAC---AGAGGCGACAUGAUUAAGUAGUA--------------- ......((.((((((.((((....)))).))).(((...((((.........))))..))).......---...))).))..............--------------- ( -16.80, z-score = 0.16, R) >droAna3.scaffold_13266 2163607 88 - 19884421 AAGCAAGUAGCUGUGGUGAUGGUUAUGGACAUGCUGAUGGCUGGCAUUGAUACUGUAAGCUAUAUUUCCUUAUAUUUUAAUCAAUUUA--------------------- .((((.((((((((....)))))))).....)))).((((((.(((.......))).)))))).........................--------------------- ( -15.50, z-score = 1.27, R) >dp4.chr3 16371846 101 - 19779522 AAGCAGGUGGCCGUAGUGAUGGCCAUGGAUAUGUUGAUGGCCGGAAUCGAUACGGUUUGUGGGAUGAUACUGUAAAG-AUCUGUAAAGAUCUCUUAAUGACA------- ..((((..(((((((.((((((((((.((....)).))))))...)))).)))))))............))))..((-((((....))))))..........------- ( -29.84, z-score = -1.93, R) >droPer1.super_2 5435478 101 - 9036312 AAGCAGGUGGCCGUAGUGAUGGCCAUGGAUAUGUUGAUGGCCGGAAUCGAUACGGUUUGUGGGAUGAUACUGUAAAG-AUCUGUAAAGAUCUCUUAAUGACA------- ..((((..(((((((.((((((((((.((....)).))))))...)))).)))))))............))))..((-((((....))))))..........------- ( -29.84, z-score = -1.93, R) >droWil1.scaffold_180697 2119634 95 + 4168966 AAACAGGUGGCUAUGGUCAUGGUCAUGGAUAUGCUAAUGGCAGGCUUAGAUACGGUUGGUUAAAAUCACUUCAAGACUGAAAGAAUUUAUUCUCA-------------- ......((((((((....))))))))(((((......(..(((.(((.((...((((......))))...))))).)))..).....)))))...-------------- ( -17.20, z-score = 0.59, R) >droVir3.scaffold_12875 6541401 105 - 20611582 AAACAGGUCGCUGUGGUUAUGGCCAUGGAUAUGUUGAUGGCUGGCUUGGAUACGGUAGGAAAUACUUCGAAUCCAGUUGGAAAUGCGUUUG-AUAUAUUUCAUGGA--- ..((((....))))........((((((((((((..(((.(..(((.((((.(((.((......))))).)))))))..).....)).)..-))))).))))))).--- ( -29.10, z-score = -1.38, R) >droGri2.scaffold_15245 17166261 100 + 18325388 AAACGGGUGGCUGUCGCUAUGGCCAUGGAUAUGUUAAUGGCUGGCAUGGAUACGGUAAAAGAUAAUACUA--CUAGUGAUGAGUCCUUAUAUAUUCAUUUCU------- ......((((((((....))))))))((((((((.((.((((.((((......((((........)))).--...))).).)))).))))))))))......------- ( -21.30, z-score = 0.45, R) >droMoj3.scaffold_6496 17351458 96 - 26866924 AAACAGGUGGCUGUGGUCAUGGCCAUGGAUAUGCUAAUGGCUGGCAUAGAUACGGUGCGAAUUGAGUUUGU-UAUCUUAAUUAGAUAUUCCUUGUAA------------ ..((((((((((((....))))))))((((((((((.....)))))))...........(((((((.....-...)))))))......)))))))..------------ ( -23.70, z-score = -0.25, R) >anoGam1.chrU 49946656 109 - 59568033 AAGCACGUUGCUGUGAUCAUGUCGCUUGAUAUGCUUAUUGCUGGAAUUGAUACGGUAUGAGCUUUGGAGGUGCAUCUUCAUUCCAAAAGUCUUCUAAAAUCAAUUUUCA .........((.((((.((((((....)))))).)))).)).(((((((((..((...((..(((((((.((......)))))))))..))..))...))))))))).. ( -22.70, z-score = 0.44, R) >triCas2.chrUn_53 436606 89 - 446649 CGCAACAUCGCCGUGACCAUGUCCAUAGACAUGAUGAUCGCAGGAAUAGACACGGUGAGGCCCAA------AAAAUCAAAAAUAAUUUAUUCGUG-------------- .((....((((((((..((((((....))))))....((....)).....)))))))).))....------........................-------------- ( -21.70, z-score = -1.86, R) >consensus AAGCAGGUGGCUGUGACCAUGGCCAUGGACAUGCUGAUGGCUGGAAUUGAUACGGUAUGAGAUAUGAU___AUAUUCGAUAUGAAUUAAUACUA_______________ .........((((((....(((((((..........))))))).......))))))..................................................... ( -9.13 = -8.73 + -0.40)
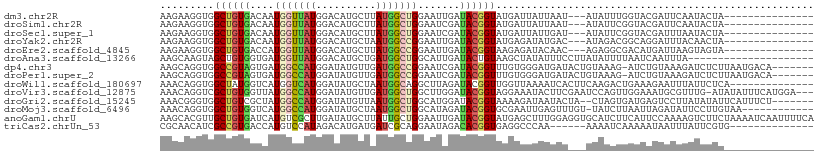


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:47 2011