| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,229,572 – 3,229,670 |
| Length | 98 |
| Max. P | 0.770038 |

| Location | 3,229,572 – 3,229,670 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.58268 |
| G+C content | 0.40636 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -10.83 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
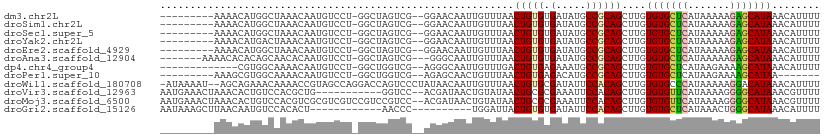
>dm3.chr2L 3229572 98 - 23011544 ---------AAAACAUGGCUAAACAAUGUCCU-GGCUAGUCG--GGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ---------...((((((.((((((((.((((-((....)))--)))...)))))))).))))))................(((((((.......)))))))........ ( -29.50, z-score = -2.88, R) >droSim1.chr2L 3168685 98 - 22036055 ---------AAAACAUGGCUAAACAAUGUCCU-GGCUAGUCG--GGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ---------...((((((.((((((((.((((-((....)))--)))...)))))))).))))))................(((((((.......)))))))........ ( -29.50, z-score = -2.88, R) >droSec1.super_5 1379050 98 - 5866729 ---------AAAACAUGGCUAAACAAUGUCCU-GGCUAGUCG--GGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ---------...((((((.((((((((.((((-((....)))--)))...)))))))).))))))................(((((((.......)))))))........ ( -29.50, z-score = -2.88, R) >droYak2.chr2L 3218791 98 - 22324452 ---------AAAACAUGACUAAACAAUGUCCU-GGCUAGUCG--GGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ---------..........((((((((.((((-((....)))--)))...)))))))).(((((........)))))....(((((((.......)))))))........ ( -27.40, z-score = -2.64, R) >droEre2.scaffold_4929 3266558 98 - 26641161 ---------AAAACAUGGCUAAACAAUGUCCU-GGCUAGUCG--GGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ---------...((((((.((((((((.((((-((....)))--)))...)))))))).))))))................(((((((.......)))))))........ ( -29.50, z-score = -2.88, R) >droAna3.scaffold_12904 38804 99 + 61084 -------AAAACACACAGCAACACAAUGUCCU-GGCUAGUCG---GGGCAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU -------....(((((((....((((((((((-((....)))---)))).)))))....)))))))...............(((((((.......)))))))........ ( -32.70, z-score = -2.97, R) >dp4.chr4_group4 5496794 94 + 6586962 -------------CGUGGCAAAACAAUGUCCU-GGCUGGUCG--AGGGCAAUUGUUUGACUGUGAGAAAUGCCGCAGCUUGUGUGCUCAUAAGAAAAGCAUAAACAUUUU -------------.((((((((((((((((((-.(.....).--))))).)))))))..(.....)...))))))..((((((....))))))................. ( -25.30, z-score = -0.58, R) >droPer1.super_10 292951 91 - 3432795 ---------AAAGCGUGGCAAAACAAUGUCCU-GGCUGGUCG--AGAGCAACUGUUUAACUGUGAGACAUGCCGCAGCUUGUGUGCUCAUAAGAAAAGCAUAA------- ---------.((((((((((......((((.(-.((.(((..--.((((....)))).))))).))))))))))).)))).((((((.........)))))).------- ( -23.40, z-score = 0.22, R) >droWil1.scaffold_180708 12050333 107 - 12563649 -AUAAAAU--AGCAGAAACAAAACCGUAGCCAGGACCAGUCCCUAUAACAAUUGUUUAACUGUGCGAUAUUCCACAGCUUGUGUGCCCAUAAAAAGGACAUAAACAUUUU -.......--.....((((((....((((...(((....))))))).....))))))..(((((.(.....)))))).((((((.((........))))))))....... ( -17.20, z-score = -0.49, R) >droVir3.scaffold_12963 6060124 97 - 20206255 AAUGAAACUAAACACUGUCCACGCUG-----------GGUCC--ACGAUAACUGUAUAACUGCGCGAAAUUCCACAGCUUGUGUGUUCAUAAAAGGGGCAUAAACGUUUU ...(((((.......(((((..((((-----------((...--.((.....((((....)))))).....)).))))(((((....)))))...))))).....))))) ( -16.00, z-score = 1.38, R) >droMoj3.scaffold_6500 25179565 108 - 32352404 AAUGAAACUAAACACUGUCCACGUCGCGUCGUCCGUCCGUCC--ACGAUAACUGUAUAACUGCGCGAAAUUCCACAGCUUGUGUGUUCAUAAAAGGGGCAUAAACGUUUU ...(((((......((((.....((((((.((....(.((..--......)).)....)).))))))......))))....(((((((.......)))))))...))))) ( -19.10, z-score = 1.37, R) >droGri2.scaffold_15126 4692004 88 - 8399593 AAUAAAGCUUAACAAUGUCCACACU------------AACCC----------UGGAUUACUGUGUGAUAUUCCACAGCUUGUGUGCUCAUAAACUGGGCAUAAACAUUUU ....(((((....((((((((((.(------------(((..----------..).))).)))).))))))....))))).((((((((.....))))))))........ ( -23.00, z-score = -2.81, R) >consensus _________AAAACAUGGCUAAACAAUGUCCU_GGCUAGUCG__AGAACAAUUGUUUAACUGUGUGAUAUGCCGCAGCUUGUGUGCUCAUAAAAAGAGCAUAAACAUUUU ...........................................................(((((.(.....))))))....(((((((.......)))))))........ (-10.83 = -10.72 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:20 2011