
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,601,331 – 14,601,470 |
| Length | 139 |
| Max. P | 0.899394 |

| Location | 14,601,331 – 14,601,470 |
|---|---|
| Length | 139 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.50627 |
| G+C content | 0.52031 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -23.75 |
| Energy contribution | -25.42 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
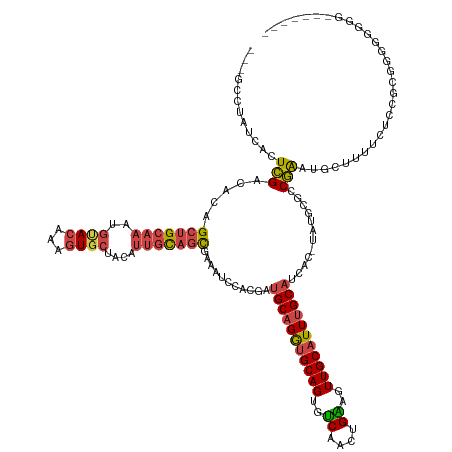
>dm3.chr2R 14601331 139 + 21146708 CACCCCCUCCCCCCCACGGAGAAAAGCAUUCGGCGAAUA-GUGAUGCAAAUGCAACUUCAGUUGACACUGCACCUGCAUCGUGGAUUUCGCUGCAAUGUAGCACUUUGUACAUUUGCAGCUGUGUCGAGUGAUAGGCAGC ..((..((((.......)))).....(((((((((((..-((((((((..((((...((....))...))))..))))))))....)))(((((((((((........)))).)))))))...))))))))...)).... ( -43.40, z-score = -1.34, R) >droSim1.chr2R 13311425 136 + 19596830 CCCCCAACCCACCCCCCGGAGAAAAGCAUUCGGCGCAUA-GUGAUGCAAAUGCAACUUCAGUUGACACUGCACCUGCAUCGUGGAUUUCGCUGCAAUGUAGCACUUUGUACAUUUGCAGCUGUGUCGAGUGAUAGGC--- ...((...((.......)).......(((((((((((..-..((((((..((((...((....))...))))..)))))).........(((((((((((........)))).))))))))))))))))))...)).--- ( -43.60, z-score = -1.70, R) >droSec1.super_1 12109114 124 + 14215200 ------------CCCCCGUAGAAAAGCAUUCGGCGCAUA-GUGAUGCAAAUGCAACUCCAGUUGACACUGCACCUGCAUCGUGGAUUUCGCUGCACUGUAGCACUUUGUACAUUUGCAGCUGUGUCGAGUGAUAGGC--- ------------..............(((((((((((..-..((((((..((((..((.....))...))))..)))))).........((((((.((((........))))..)))))))))))))))))......--- ( -38.80, z-score = -0.55, R) >droYak2.chr2R 6540709 131 - 21139217 ------CCCUCCCCCGCCGAGUGAAGCAUUCGGGGAAAAAGCGUUGCAAAUGCAACUUCAGUUGACACUGCACCUGCAGCAAGAAUUUCGCUGCAAUGUGGCACUUUGUACAUUUGCAGCUGUGUCGAGUGAUAGGC--- ------.(((...((.(((((((...))))))))).......(((((....))))).(((.((((((((((....)))...........(((((((((((.(.....))))).))))))).))))))).))).))).--- ( -46.40, z-score = -1.83, R) >droEre2.scaffold_4845 8792556 129 + 22589142 -------CCCCCUCCGCCAGGAGAAGCGUUCGGGGAGAA-GUGAUGCAAAUGCAACUCCAGUUGACACUGCACCUGCAGCAAGGAUUUCGCUGCAAUGUAGCACUUUGUGCACUUGCAGCUGUGUCGAGUGAUAGGC--- -------((((((((....))))........))))....-....(((....)))....((.(((((((....(((......))).....(((((((.((.((((...))))))))))))).))))))).))......--- ( -44.40, z-score = -0.47, R) >droAna3.scaffold_13266 2108002 117 - 19884421 ----------------CAAAAACAGGGGCCUGCAUCCAAGGGGUAGCCAAUGCAAAUGCAACUGCCUCUGCCUCUGC--CACAGAUGCCAAAAGGAAAAAGAA--UACUAGAAUUGCGGCAGUGGCAACUGAUAGGC--- ----------------...........(((((((....((((((((....(((....))).))))))))(((.((((--(.(((...((....))....((..--..))....))).))))).)))...)).)))))--- ( -36.30, z-score = -0.68, R) >consensus _______CCCCCCCCCCGGAGAAAAGCAUUCGGCGAAAA_GUGAUGCAAAUGCAACUUCAGUUGACACUGCACCUGCAUCAUGGAUUUCGCUGCAAUGUAGCACUUUGUACAUUUGCAGCUGUGUCGAGUGAUAGGC___ ..........................(((((((.........((((((..((((...((....))...))))..)))))).........(((((((((((........)))).)))))))....)))))))......... (-23.75 = -25.42 + 1.67)



| Location | 14,601,331 – 14,601,470 |
|---|---|
| Length | 139 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.50627 |
| G+C content | 0.52031 |
| Mean single sequence MFE | -45.64 |
| Consensus MFE | -25.58 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14601331 139 - 21146708 GCUGCCUAUCACUCGACACAGCUGCAAAUGUACAAAGUGCUACAUUGCAGCGAAAUCCACGAUGCAGGUGCAGUGUCAACUGAAGUUGCAUUUGCAUCAC-UAUUCGCCGAAUGCUUUUCUCCGUGGGGGGGAGGGGGUG (((.(((....(((......(((((((.((((........)))))))))))....((((((((((((((((((..((....))..)))))))))))))..-........(((.....)))...)))))))).))).))). ( -52.40, z-score = -2.43, R) >droSim1.chr2R 13311425 136 - 19596830 ---GCCUAUCACUCGACACAGCUGCAAAUGUACAAAGUGCUACAUUGCAGCGAAAUCCACGAUGCAGGUGCAGUGUCAACUGAAGUUGCAUUUGCAUCAC-UAUGCGCCGAAUGCUUUUCUCCGGGGGGUGGGUUGGGGG ---........((((((...(((((((.((((........))))))))))).....(((((((((((((((((..((....))..)))))))))))))..-....(.(((............))).).)))))))))).. ( -52.10, z-score = -2.49, R) >droSec1.super_1 12109114 124 - 14215200 ---GCCUAUCACUCGACACAGCUGCAAAUGUACAAAGUGCUACAGUGCAGCGAAAUCCACGAUGCAGGUGCAGUGUCAACUGGAGUUGCAUUUGCAUCAC-UAUGCGCCGAAUGCUUUUCUACGGGGG------------ ---........((((.....((((((...((((...)))).....))))))((((.....(((((((((((((..((....))..)))))))))))))..-...(((.....))).))))..))))..------------ ( -40.10, z-score = -0.86, R) >droYak2.chr2R 6540709 131 + 21139217 ---GCCUAUCACUCGACACAGCUGCAAAUGUACAAAGUGCCACAUUGCAGCGAAAUUCUUGCUGCAGGUGCAGUGUCAACUGAAGUUGCAUUUGCAACGCUUUUUCCCCGAAUGCUUCACUCGGCGGGGGAGGG------ ---.....(((...(((((.(.(((....))))...(..((....((((((((.....))))))))))..).)))))...))).(((((....))))).((((..((((((.((...)).)))).))..)))).------ ( -46.60, z-score = -1.85, R) >droEre2.scaffold_4845 8792556 129 - 22589142 ---GCCUAUCACUCGACACAGCUGCAAGUGCACAAAGUGCUACAUUGCAGCGAAAUCCUUGCUGCAGGUGCAGUGUCAACUGGAGUUGCAUUUGCAUCAC-UUCUCCCCGAACGCUUCUCCUGGCGGAGGGGG------- ---...........(((...((((((.((((((...)))).)).(((((((((.....))))))))).)))))))))....((((((((....)))..))-))).((((...((((......))))..)))).------- ( -48.00, z-score = -1.79, R) >droAna3.scaffold_13266 2108002 117 + 19884421 ---GCCUAUCAGUUGCCACUGCCGCAAUUCUAGUA--UUCUUUUUCCUUUUGGCAUCUGUG--GCAGAGGCAGAGGCAGUUGCAUUUGCAUUGGCUACCCCUUGGAUGCAGGCCCCUGUUUUUG---------------- ---(((((((..(((((.((((((((((.((((..--............)))).)).))))--)))).))))).(((...(((....)))...)))........)))..))))...........---------------- ( -34.64, z-score = 0.11, R) >consensus ___GCCUAUCACUCGACACAGCUGCAAAUGUACAAAGUGCUACAUUGCAGCGAAAUCCACGAUGCAGGUGCAGUGUCAACUGAAGUUGCAUUUGCAUCAC_UAUGCGCCGAAUGCUUUUCUCCGCGGGGGGGG_______ ............(((.....(((((((..((((...))))....))))))).........(.(((((((((((..((....))..))))))))))).)..........)))............................. (-25.58 = -26.03 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:42 2011