
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,574,679 – 14,574,770 |
| Length | 91 |
| Max. P | 0.887476 |

| Location | 14,574,679 – 14,574,770 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
| Shannon entropy | 0.32837 |
| G+C content | 0.51650 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -23.13 |
| Energy contribution | -24.69 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
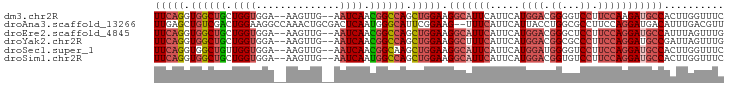
>dm3.chr2R 14574679 91 + 21146708 UUCAGGUGGCUGCUGGUGGA--AAGUUG--AAUCAACGGCCAGCUGGAAGGCAUUCAUUCAUGGACGGGGUCCUUCCAAGAUGCCACUUGGUUUC .(((((((((.((((((...--..((((--...)))).))))))(((((((.((((.((....))..)))))))))))....))))))))).... ( -36.20, z-score = -2.55, R) >droAna3.scaffold_13266 15090131 93 - 19884421 UUGAGCUGUCGACUGGAAGGCCAAACUGCGACUCAAUGGGCAUUCGGAAG--UUUCAUUCAUUACCUGGCGCCUUCCAGGAUGACAUUUGACGUU (..((.(((((.(((((((((......((.(...((((((....(....)--.....))))))...).)))))))))))..)))))))..).... ( -26.40, z-score = -0.71, R) >droEre2.scaffold_4845 8764418 91 + 22589142 UUCAGGUGGCUGCUGGUGGA--AAGUUG--AAUCAACGGCCAGCUGGAAGGCAUUCAUUCAUGGACGGGCUCCUUCCAGGAUGCCAUUUAGUUUG (((((.((((((.((((...--......--.)))).)))))).))))).(((((((.....((((.((...)).))))))))))).......... ( -33.80, z-score = -1.93, R) >droYak2.chr2R 6513170 91 - 21139217 UUCAGGUGGCUGCUGGUGGA--AAGUUG--AAUCAACGGCCAGCUGGAAGGCUUUCAUUCAUGGACGGCGCCCUUCCAGGAUGCCGAUUAGUUUG (((((.((((((.((((...--......--.)))).)))))).)))))(((((((((....))))(((((.((.....)).)))))...))))). ( -31.00, z-score = -0.66, R) >droSec1.super_1 12082208 91 + 14215200 UUCAGGUGGCUGUUGGUGGA--AAGUUG--AAUCAACGGCAAGCUGGAAGGCAUUCAUUCAUGGAUGGGGUCCUUCCAGGAUGCCACUUGGUUUC .((((((((((((((((...--......--.)))))))))...((((((((..((((((....))))))..)))))))).....))))))).... ( -38.10, z-score = -3.79, R) >droSim1.chr2R 13280204 91 + 19596830 UUCAGGUGGCUGCUGGUGGA--AAGUUG--AAUCAAUGGCCAGCUGGAAGGCAUUCAUUCAUGGACGGUGUCCUUCCAGGAUGCCACUUGGUUUC .(((((((((.((((((...--..((((--...)))).))))))(((((((((((..((....)).)))).)))))))....))))))))).... ( -35.00, z-score = -2.14, R) >consensus UUCAGGUGGCUGCUGGUGGA__AAGUUG__AAUCAACGGCCAGCUGGAAGGCAUUCAUUCAUGGACGGGGUCCUUCCAGGAUGCCACUUGGUUUC (((((.((((((.((((..............)))).)))))).))))).(((((((.....((((.((...)).))))))))))).......... (-23.13 = -24.69 + 1.56)


| Location | 14,574,679 – 14,574,770 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Shannon entropy | 0.32837 |
| G+C content | 0.51650 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -17.56 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
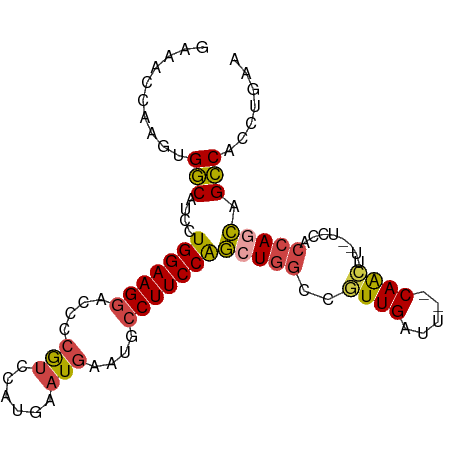
>dm3.chr2R 14574679 91 - 21146708 GAAACCAAGUGGCAUCUUGGAAGGACCCCGUCCAUGAAUGAAUGCCUUCCAGCUGGCCGUUGAUU--CAACUU--UCCACCAGCAGCCACCUGAA .....((.(((((....(((((((....(((......)))....)))))))(((((..((((...--))))..--....))))).))))).)).. ( -29.70, z-score = -2.84, R) >droAna3.scaffold_13266 15090131 93 + 19884421 AACGUCAAAUGUCAUCCUGGAAGGCGCCAGGUAAUGAAUGAAA--CUUCCGAAUGCCCAUUGAGUCGCAGUUUGGCCUUCCAGUCGACAGCUCAA .........((((...(((((((((..((.....))....(((--((..(((...(.....)..))).))))).)))))))))..))))...... ( -23.90, z-score = -0.52, R) >droEre2.scaffold_4845 8764418 91 - 22589142 CAAACUAAAUGGCAUCCUGGAAGGAGCCCGUCCAUGAAUGAAUGCCUUCCAGCUGGCCGUUGAUU--CAACUU--UCCACCAGCAGCCACCUGAA .........((((....(((((((....(((......)))....)))))))(((((..((((...--))))..--....))))).))))...... ( -25.30, z-score = -1.44, R) >droYak2.chr2R 6513170 91 + 21139217 CAAACUAAUCGGCAUCCUGGAAGGGCGCCGUCCAUGAAUGAAAGCCUUCCAGCUGGCCGUUGAUU--CAACUU--UCCACCAGCAGCCACCUGAA ..........(((....(((((((.(..(((......)))...))))))))(((((..((((...--))))..--....))))).)))....... ( -24.50, z-score = -0.48, R) >droSec1.super_1 12082208 91 - 14215200 GAAACCAAGUGGCAUCCUGGAAGGACCCCAUCCAUGAAUGAAUGCCUUCCAGCUUGCCGUUGAUU--CAACUU--UCCACCAACAGCCACCUGAA .....((.(((((...((((((((....(((......)))....))))))))......((((...--......--.....)))).))))).)).. ( -26.24, z-score = -3.14, R) >droSim1.chr2R 13280204 91 - 19596830 GAAACCAAGUGGCAUCCUGGAAGGACACCGUCCAUGAAUGAAUGCCUUCCAGCUGGCCAUUGAUU--CAACUU--UCCACCAGCAGCCACCUGAA .....((.(((((...((((..((((...)))).(((((.((((((........)).)))).)))--))....--....))))..))))).)).. ( -30.60, z-score = -3.27, R) >consensus GAAACCAAGUGGCAUCCUGGAAGGACCCCGUCCAUGAAUGAAUGCCUUCCAGCUGGCCGUUGAUU__CAACUU__UCCACCAGCAGCCACCUGAA ..........(((....(((((((....(((......)))....)))))))(((((..((((.....))))........))))).)))....... (-17.56 = -17.70 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:38 2011