
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,561,510 – 14,561,609 |
| Length | 99 |
| Max. P | 0.753132 |

| Location | 14,561,510 – 14,561,609 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 56.06 |
| Shannon entropy | 0.89399 |
| G+C content | 0.36589 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -4.96 |
| Energy contribution | -6.66 |
| Covariance contribution | 1.71 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
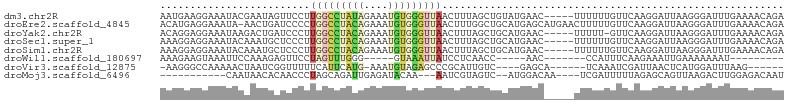
>dm3.chr2R 14561510 99 + 21146708 AAUGAAGGAAAUACGAAUAGUUCCUUGGCCUAUAGAAAUGUGGGUUAACUUUAGCUGUAUGAAC-----UUUUUUGUUCAAGGAUUAAGGGAUUUGAAAACAGA ....((((((.((....)).))))))((((((((....))))))))..((((((((...(((((-----......))))).)).)))))).............. ( -23.30, z-score = -2.08, R) >droEre2.scaffold_4845 8751056 103 + 22589142 ACAUGAGGAAAUA-AACUGAUCCCCUGGCCUACAGAAAUGUGGGUUAACUUUGGCUGCAUGAGCAUGAACUUUUUGUUCAAGGAUUAAGGGAUUUGAAAACAGA .............-..(.((((((.(((((((((....))))))))).((((.(((.....)))..((((.....)))))))).....)))))).)........ ( -27.90, z-score = -2.00, R) >droYak2.chr2R 6498642 98 - 21139217 ACAGGAGGAAAUAAGACUGAUCCCUUGGCCUACAGAAAUGUGGGUUAACUUUAGCUGCAUGAAC-----UUUUU-GUUCAAGGAUUAAGGGAUUUGAAAACAGA ................(.((((((((((((((((....)))))))..............(((((-----.....-))))).....))))))))).)........ ( -27.10, z-score = -2.31, R) >droSec1.super_1 12068989 99 + 14215200 AAAGGAGGAAAUACAAAUGCUCCCUUGGCCUACAGAAAUGUGGGUUAACUUUAGCUGCAUGAAC-----UUUUUUGUUCAAGGAUUAAGGGAUUUGAAAACAGA ...((((.(........).)))).((((((((((....))))))))))((((((((...(((((-----......))))).)).)))))).............. ( -27.00, z-score = -2.14, R) >droSim1.chr2R 13266600 99 + 19596830 AAAGGAGGAAAUACAAAUGCUCCCUUGGCCUACAGAAAUGUGGGUUAACUUUAGCUGCAUGAAC-----UUUUUUGUUCAAGGAUUAAGGGAUUUGAAAACAGA ...((((.(........).)))).((((((((((....))))))))))((((((((...(((((-----......))))).)).)))))).............. ( -27.00, z-score = -2.14, R) >droWil1.scaffold_180697 1979301 78 + 4168966 AAAGAAGUAAAUUCCAAAGAGUUCCUAGUUUGGG-----GUAAAUUAUCCUCAACC-----AAC-------CCAUUUCAAGAAAUUGAAAAAAAU--------- .........(((((....)))))....(.(((((-----(........)))))).)-----...-------...((((((....)))))).....--------- ( -12.50, z-score = -1.15, R) >droVir3.scaffold_12875 6430511 86 - 20611582 -AAGGGCCAAAAACUAAUCGGUUUUUCAUUCAUG-AAAUGUAGAGCCCGCAUUGUC----GAGCA------UCAAAUCGAUUAACUCAUGGAUUUAAG------ -(((((((...........))))))).(((((((-((((((.......)))))(((----((...------.....)))))....)))))))).....------ ( -15.50, z-score = -0.29, R) >droMoj3.scaffold_6496 17237072 84 - 26866924 -----------CAAUAACACAACCCUAGCAGAUUGAGAUACAA---AAUCGUAGUC--AUGGACAA----UCGAUUUUUAGAGCAGUUAAGACUUGGAGACAAU -----------...................(((((...((.((---(((((..(((--...)))..----.)))))))))...))))).....(((....))). ( -12.70, z-score = -0.91, R) >consensus AAAGGAGGAAAUACAAAUGAUCCCUUGGCCUACAGAAAUGUGGGUUAACUUUAGCUGCAUGAAC_____UUUUUUGUUCAAGGAUUAAGGGAUUUGAAAACAGA .........................(((((((((....)))))))))......................................................... ( -4.96 = -6.66 + 1.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:37 2011