| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,558,276 – 14,558,381 |
| Length | 105 |
| Max. P | 0.940165 |

| Location | 14,558,276 – 14,558,381 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.27946 |
| G+C content | 0.37123 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.51 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
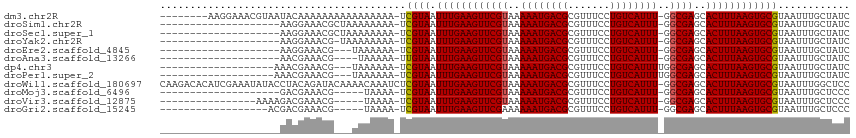
>dm3.chr2R 14558276 105 + 21146708 --------AAGGAAACGUAAUACAAAAAAAAAAAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------.(((((((((.....................-(((.....))).((.(((....))))))))))))))......(-(((((((((.....)))).....)))))).. ( -22.20, z-score = -1.92, R) >droSim1.chr2R 13263414 93 + 19596830 --------------------AAGGAAACGCUAAAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------------------.(((((((((.........-(((.....))).((.(((....))))))))))))))......(-(((((((((.....)))).....)))))).. ( -24.10, z-score = -2.38, R) >droSec1.super_1 12065821 93 + 14215200 --------------------AAGGAAACGCUAAAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------------------.(((((((((.........-(((.....))).((.(((....))))))))))))))......(-(((((((((.....)))).....)))))).. ( -24.10, z-score = -2.38, R) >droYak2.chr2R 6495309 92 - 21139217 --------------------AAGGAAACG-UAAAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------------------..(....)(-((((.....-.((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))...))))).... ( -22.70, z-score = -2.12, R) >droEre2.scaffold_4845 8747576 90 + 22589142 --------------------AAGGAAACG---UAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------------------..(....)(---((((...-.((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))...))))).... ( -23.90, z-score = -2.55, R) >droAna3.scaffold_13266 15070727 89 - 19884421 --------------------AACGAAACG----UAAAAA-UUGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC --------------------........(----((((..-.((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))...))))).... ( -21.20, z-score = -1.56, R) >dp4.chr3 16269994 92 - 19779522 -------------------AAACGAAACG---UAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUUUGGCGAGCACUUUAAGUGCGUAAUUUGCUAUC -------------------.........(---((((...-.((((.((((((((((((.(((((((((.......))))))))).))))..))))))))))))...))))).... ( -24.60, z-score = -2.60, R) >droPer1.super_2 5338674 92 - 9036312 -------------------AAACGAAACG---UAAAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUUUGGCGAGCACUUUAAGUGCGUAAUUUGCUAUC -------------------.........(---((((...-.((((.((((((((((((.(((((((((.......))))))))).))))..))))))))))))...))))).... ( -24.60, z-score = -2.60, R) >droWil1.scaffold_180697 1972700 114 + 4168966 CAAGACACAUCGAAAUAUACCUACAGAUACAAAACAAAUCUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGGCUCC ..................................(((((..((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))..)))))..... ( -24.20, z-score = -1.34, R) >droMoj3.scaffold_6496 17230905 88 - 26866924 --------------------GACGAAACG-----UAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUCCC --------------------........(-----((((.-.((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))...))))).... ( -22.60, z-score = -1.93, R) >droVir3.scaffold_12875 6425312 92 - 20611582 ----------------AAAAGACGAAACG-----UAAAA-UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUCCC ----------------............(-----((((.-.((((.((((((((((((..((((((((.......))))))))-.))))..))))))))))))...))))).... ( -22.60, z-score = -1.84, R) >droGri2.scaffold_15245 4166033 90 - 18325388 ------------------ACGACGAAACG-----UAAAA-UCGUAAUUUGAAGUUCGAAAAAAUGACGCGUUUCCUGUCAUUU-GGCGAGCACUUUAAGUGCGUAAUUUGCUCCC ------------------..........(-----((((.-.((((.(((((((((((...((((((((.......))))))))-..)))..))))))))))))...))))).... ( -22.50, z-score = -1.42, R) >consensus ____________________AACGAAACG___AAAAAAA_UCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUU_GGCGAGCACUUUAAGUGCGUAAUUUGCUAUC .........................................((((.((((((((((((..((((((((.......))))))))..))))..))))))))))))............ (-18.50 = -18.51 + 0.01)


| Location | 14,558,276 – 14,558,381 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.27946 |
| G+C content | 0.37123 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14558276 105 - 21146708 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUUUUUUUUUUUGUAUUACGUUUCCUU-------- ....(((((....((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.............))))).............-------- ( -18.90, z-score = -1.65, R) >droSim1.chr2R 13263414 93 - 19596830 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUUUAGCGUUUCCUU-------------------- ((..((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.........))...))...-------------------- ( -19.00, z-score = -1.76, R) >droSec1.super_1 12065821 93 - 14215200 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUUUAGCGUUUCCUU-------------------- ((..((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.........))...))...-------------------- ( -19.00, z-score = -1.76, R) >droYak2.chr2R 6495309 92 + 21139217 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUUUA-CGUUUCCUU-------------------- ((.(((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.........-.)))))...-------------------- ( -17.10, z-score = -1.64, R) >droEre2.scaffold_4845 8747576 90 - 22589142 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUA---CGUUUCCUU-------------------- ((.(((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.......---.)))))...-------------------- ( -17.10, z-score = -1.68, R) >droAna3.scaffold_13266 15070727 89 + 19884421 GAUAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACAA-UUUUUA----CGUUUCGUU-------------------- ((.(((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-......----.)))))...-------------------- ( -18.00, z-score = -1.86, R) >dp4.chr3 16269994 92 + 19779522 GAUAGCAAAUUACGCACUUAAAGUGCUCGCCAAAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUA---CGUUUCGUUU------------------- ((.(((.......((((.....))))(((..((((((((.(....)..))))))))..)))..............-.......---.)))))....------------------- ( -20.00, z-score = -2.00, R) >droPer1.super_2 5338674 92 + 9036312 GAUAGCAAAUUACGCACUUAAAGUGCUCGCCAAAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUUUA---CGUUUCGUUU------------------- ((.(((.......((((.....))))(((..((((((((.(....)..))))))))..)))..............-.......---.)))))....------------------- ( -20.00, z-score = -2.00, R) >droWil1.scaffold_180697 1972700 114 - 4168966 GGAGCCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAGAUUUGUUUUGUAUCUGUAGGUAUAUUUCGAUGUGUCUUG ((((.........((((.....))))(((..-(((((((.(....)..)))))))...))).))))......(((((((((...((((((....))))))...)))...)))))) ( -28.20, z-score = -1.61, R) >droMoj3.scaffold_6496 17230905 88 + 26866924 GGGAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUA-----CGUUUCGUC-------------------- .(((((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.....-----.)))))...-------------------- ( -21.30, z-score = -2.10, R) >droVir3.scaffold_12875 6425312 92 + 20611582 GGGAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA-UUUUA-----CGUUUCGUCUUUU---------------- .(((((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.....-----.))))).......---------------- ( -21.30, z-score = -1.94, R) >droGri2.scaffold_15245 4166033 90 + 18325388 GGGAGCAAAUUACGCACUUAAAGUGCUCGCC-AAAUGACAGGAAACGCGUCAUUUUUUCGAACUUCAAAUUACGA-UUUUA-----CGUUUCGUCGU------------------ .(((((.......((((.....))))(((..-(((((((.(....)..)))))))...)))..............-.....-----.))))).....------------------ ( -21.30, z-score = -1.36, R) >consensus GAUAGCAAAUUACGCACUUAAAGUGCUCGCC_AAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGA_UUUUUUU___CGUUUCGUU____________________ ((.(((.......((((.....))))(((...(((((((.(....)..)))))))...)))..........................)))))....................... (-17.18 = -17.07 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:36 2011