
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,520,381 – 14,520,471 |
| Length | 90 |
| Max. P | 0.859908 |

| Location | 14,520,381 – 14,520,471 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Shannon entropy | 0.55627 |
| G+C content | 0.60504 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -14.23 |
| Energy contribution | -13.46 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
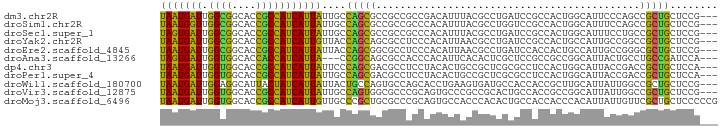
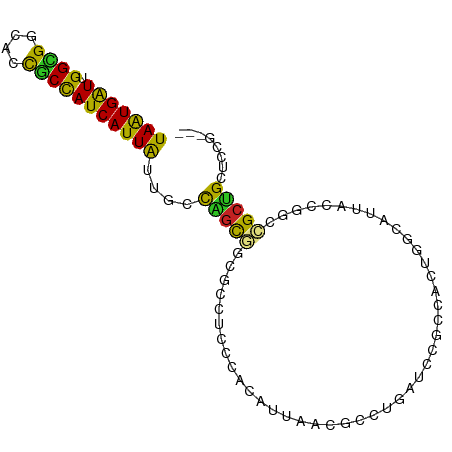
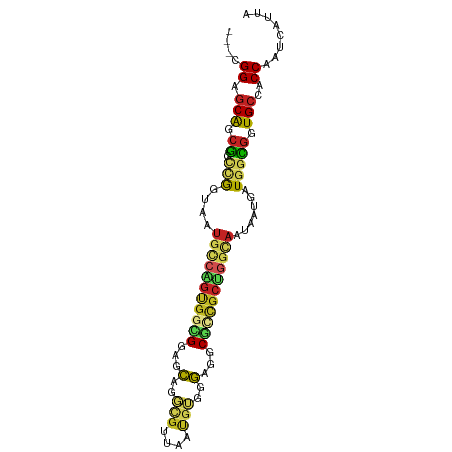
>dm3.chr2R 14520381 90 + 21146708 UAAUGAUUGGCGGCACCGCCAUCAUUAUUGCCAGCGCCGCCGCCGACAUUUACGCCUGAUCCGCCACUGGCAUUCCCAGCCGCUGCUCCG--- .((((.((((((((..(((...((....))...)))..)))))))))))).......((.(.((..((((.....))))..)).).))..--- ( -27.10, z-score = -0.96, R) >droSim1.chr2R 13224776 90 + 19596830 UAAUGGUUGGCGGCACCGCCAUCAUUAUUGCCAGCGCCGCCGCCCACAUUUACGCCUGGUCCGCCACUGGCAUUUCCAGCCGCUGCUCCG--- ...(((.(((((((.(.(.((.......)).).).))))))).)))......((.((((...(((...)))....)))).))........--- ( -28.10, z-score = -0.08, R) >droSec1.super_1 12028102 90 + 14215200 UAGUGAUUGGCGGCACCGCCAUCAUUAUUGCCAGCGCCGCCGCCCACAUUUACGCCUGAUCCGCCACUGGCAUUUCCUGCCGCUGCUCCG--- ..(((..(((((((.(.(.((.......)).).).)))))))..)))...............((...(((((.....)))))..))....--- ( -25.60, z-score = -0.47, R) >droYak2.chr2R 6455878 90 - 21139217 UAAUGAUUGGCGGCACCGCCAUCAUUGUUACCAGCAGCGCCUCCCACAUUAACGCCUGAUCCGCCACUGCCAUUGCCGGCCGCUGCUCCG--- (((((((.((((....))))))))))).....(((((((..............((.......))....(((......))))))))))...--- ( -28.00, z-score = -1.81, R) >droEre2.scaffold_4845 8708179 90 + 22589142 UAAUGAUUGGCGGCACCGCCAUCAUUAUUACCAGCGGCGCCUCCCACAUUAACGCCUGAUCCACCACUGCCAUUGCCGGGCGCUGCUCCG--- (((((((.((((....))))))))))).....((((((((((....((.....((.((......))..))...))..))))))))))...--- ( -30.90, z-score = -2.49, R) >droAna3.scaffold_13266 6887337 87 + 19884421 UAGUGAUUGGUGGCACCACCAUCAUUA---CCGGCAGCGCCACCCACAUUCACACUCGCUCCGCCGCCGGCAUUACUGCCUGCCGAUCCA--- (((((((.((((....)))))))))))---.((((((((.................))))..)))).(((((........))))).....--- ( -27.23, z-score = -2.09, R) >dp4.chr3 10581219 90 + 19779522 UAAUGAUUGGUGGCACCGCCAUCAUUAUUCCCAGCGACGCCUCCUACACUGCCGCUCGCGCCUCCACUGGCAUUACCGACCGCUGCUCCA--- (((((((.((((....)))))))))))....(((((..............((.....))(((......))).........))))).....--- ( -25.00, z-score = -1.87, R) >droPer1.super_4 5909730 90 + 7162766 UAAUGAUUGGUGGCACCGCCAUCAUUAUUGCCAGCGACGCCUCCUACACUGCCGCUCGCGCCUCCACUGGCAUUACCGACCGCUGCUCCA--- (((((((.((((....)))))))))))..((.((((..............((.....))(((......))).........))))))....--- ( -25.10, z-score = -1.29, R) >droWil1.scaffold_180700 1808756 90 - 6630534 UAAUGAUUGGAGGCAUUACUAUCAUUAUUACUGCCAGUGCCAGCACCUGAAGUGAUGCCACCACCGCUUGCAUUAUUGGCCGCUGCUCCG--- ........(((((((.((.((....)).)).)))(((((((((....(((((((.((....)).))))).))...)))).))))))))).--- ( -21.40, z-score = -0.01, R) >droVir3.scaffold_12875 9465617 90 - 20611582 UAAUGAUUGGUGGCACCGCCAUCAUUAUUGCCAGUGGCGCCCGCAGUGCCCGCCGCACUGCCACCGCCGGCAUUAUUGGCCGCUGCUCCG--- (((((((.((((....)))))))))))..((.(((((((...(((((((.....)))))))...))))(((.......))))))))....--- ( -38.80, z-score = -2.04, R) >droMoj3.scaffold_6496 15781124 93 + 26866924 UAAUGAUUGGUGGCACCGCCAUCAUUGUUGCCCGCUGCGCCCGCAGUGCCACCCACACUGCCACCACCCACAUUAUUGUUCGCUGCUCCCCCG (((((((.((((....)))))))))))......((.(((...((((((.......))))))........(((....))).))).))....... ( -25.60, z-score = -1.83, R) >consensus UAAUGAUUGGCGGCACCGCCAUCAUUAUUGCCAGCGGCGCCUCCCACAUUAACGCCUGAUCCGCCACUGGCAUUACCGGCCGCUGCUCCG___ (((((((.((((....)))))))))))....(((((............................................)))))........ (-14.23 = -13.46 + -0.76)
| Location | 14,520,381 – 14,520,471 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.72 |
| Shannon entropy | 0.55627 |
| G+C content | 0.60504 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.661875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
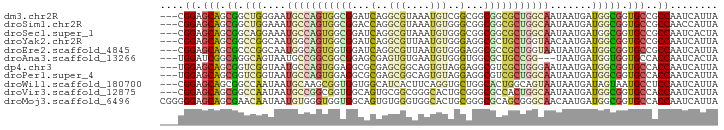
>dm3.chr2R 14520381 90 - 21146708 ---CGGAGCAGCGGCUGGGAAUGCCAGUGGCGGAUCAGGCGUAAAUGUCGGCGGCGGCGCUGGCAAUAAUGAUGGCGGUGCCGCCAAUCAUUA ---..((((.((.(((((.....))))).))......((((....)))).))((((((((((.((.......)).))))))))))..)).... ( -40.40, z-score = -2.00, R) >droSim1.chr2R 13224776 90 - 19596830 ---CGGAGCAGCGGCUGGAAAUGCCAGUGGCGGACCAGGCGUAAAUGUGGGCGGCGGCGCUGGCAAUAAUGAUGGCGGUGCCGCCAACCAUUA ---.......(((.((((...((((...))))..)))).)))....((((..((((((((((.((.......)).))))))))))..)))).. ( -43.80, z-score = -2.57, R) >droSec1.super_1 12028102 90 - 14215200 ---CGGAGCAGCGGCAGGAAAUGCCAGUGGCGGAUCAGGCGUAAAUGUGGGCGGCGGCGCUGGCAAUAAUGAUGGCGGUGCCGCCAAUCACUA ---....((.((((((.....)))).)).))...............((((..((((((((((.((.......)).))))))))))..)))).. ( -37.30, z-score = -1.35, R) >droYak2.chr2R 6455878 90 + 21139217 ---CGGAGCAGCGGCCGGCAAUGGCAGUGGCGGAUCAGGCGUUAAUGUGGGAGGCGCUGCUGGUAACAAUGAUGGCGGUGCCGCCAAUCAUUA ---...(((((((.((.....(.(((.(((((.......))))).))).)..)))))))))......((((((((((....)))).)))))). ( -34.80, z-score = -0.63, R) >droEre2.scaffold_4845 8708179 90 - 22589142 ---CGGAGCAGCGCCCGGCAAUGGCAGUGGUGGAUCAGGCGUUAAUGUGGGAGGCGCCGCUGGUAAUAAUGAUGGCGGUGCCGCCAAUCAUUA ---(((.((...)))))....((.((((((((..((..(((....)))..))..)))))))).)).(((((((((((....)))).))))))) ( -35.60, z-score = -0.75, R) >droAna3.scaffold_13266 6887337 87 - 19884421 ---UGGAUCGGCAGGCAGUAAUGCCGGCGGCGGAGCGAGUGUGAAUGUGGGUGGCGCUGCCGG---UAAUGAUGGUGGUGCCACCAAUCACUA ---....((.((.((((....))))....)).))......((((.....((((((((..(((.---......)))..))))))))..)))).. ( -39.40, z-score = -2.56, R) >dp4.chr3 10581219 90 - 19779522 ---UGGAGCAGCGGUCGGUAAUGCCAGUGGAGGCGCGAGCGGCAGUGUAGGAGGCGUCGCUGGGAAUAAUGAUGGCGGUGCCACCAAUCAUUA ---..(((((...((((.(..((((......))))..).))))..))).((.(((..(((((..........)))))..))).))..)).... ( -29.30, z-score = 0.17, R) >droPer1.super_4 5909730 90 - 7162766 ---UGGAGCAGCGGUCGGUAAUGCCAGUGGAGGCGCGAGCGGCAGUGUAGGAGGCGUCGCUGGCAAUAAUGAUGGCGGUGCCACCAAUCAUUA ---(((.((.((.((((.((.(((((((((..((....)).((..........)).))))))))).)).)))).)).)).))).......... ( -32.40, z-score = -0.49, R) >droWil1.scaffold_180700 1808756 90 + 6630534 ---CGGAGCAGCGGCCAAUAAUGCAAGCGGUGGUGGCAUCACUUCAGGUGCUGGCACUGGCAGUAAUAAUGAUAGUAAUGCCUCCAAUCAUUA ---.(((((((((........)))...(((((.(((((((......))))))).)))))...................)).))))........ ( -26.10, z-score = 0.21, R) >droVir3.scaffold_12875 9465617 90 + 20611582 ---CGGAGCAGCGGCCAAUAAUGCCGGCGGUGGCAGUGCGGCGGGCACUGCGGGCGCCACUGGCAAUAAUGAUGGCGGUGCCACCAAUCAUUA ---.((.(((.(.((((.((.((((((.(((((((((((.....)))))))...)))).)))))).))....)))).))))..))........ ( -40.20, z-score = -0.77, R) >droMoj3.scaffold_6496 15781124 93 - 26866924 CGGGGGAGCAGCGAACAAUAAUGUGGGUGGUGGCAGUGUGGGUGGCACUGCGGGCGCAGCGGGCAACAAUGAUGGCGGUGCCACCAAUCAUUA .......((.((..(((....)))..))....))((((..(((((((((((.(...((...(....)..)).).)))))))))))...)))). ( -34.30, z-score = -1.10, R) >consensus ___CGGAGCAGCGGCCGGUAAUGCCAGUGGCGGAGCAGGCGUUAAUGUGGGAGGCGCCGCUGGCAAUAAUGAUGGCGGUGCCACCAAUCAUUA ....((.(((.((.(((....(((((((((((...(..(((....)))..)...))))))))))).......))))).)))..))........ (-18.53 = -18.55 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:28 2011