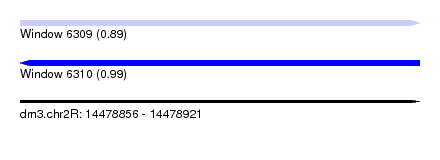
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,478,856 – 14,478,921 |
| Length | 65 |
| Max. P | 0.994847 |

| Location | 14,478,856 – 14,478,921 |
|---|---|
| Length | 65 |
| Sequences | 4 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 52.56 |
| Shannon entropy | 0.79853 |
| G+C content | 0.47333 |
| Mean single sequence MFE | -14.73 |
| Consensus MFE | -5.90 |
| Energy contribution | -6.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
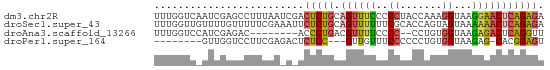
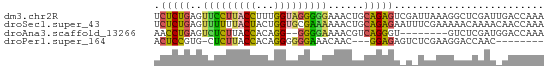
>dm3.chr2R 14478856 65 + 21146708 UUUGGUCAAUCGAGCCUUUAAUCGACUCUGCAGUUUCCCCCUACCAAAGGUAAGGAACUCAGAGA .........((((........))))(((((.((((((..(((.....)))...))))))))))). ( -17.90, z-score = -1.92, R) >droSec1.super_43 56432 65 + 327307 UUUGGUUGUUUUGUUUUUCGAAAUUCUCUGCAGUUUUUUCGCACCAGUAGUAAAAAACUCAGAGA .......((((((.....))))))((((((.((((((((.((.......)))))))))))))))) ( -15.10, z-score = -2.19, R) >droAna3.scaffold_13266 16962352 55 - 19884421 UUUGGUCCAUCGAGAC--------ACCCUGACGUUUUCCCC--CCUGUGGUAAGAGACUCAGGUU ....(((......)))--------..(((((.(((((...(--(....))...)))))))))).. ( -12.20, z-score = 0.22, R) >droPer1.super_164 85937 53 + 94152 --------GUUGGUCCUUCGAGACUCUCC---GUUGUUUCCCCCCUGUGGUAAGAG-CACGGAGU --------...((((......))))((((---((.((((...((....))...)))-))))))). ( -13.70, z-score = -0.42, R) >consensus UUUGGUC_AUCGAGCCUUCGA_A_ACUCUGCAGUUUUCCCCCACCAGUGGUAAGAAACUCAGAGA .........................(((((.((((((..((.......))...))))))))))). ( -5.90 = -6.15 + 0.25)
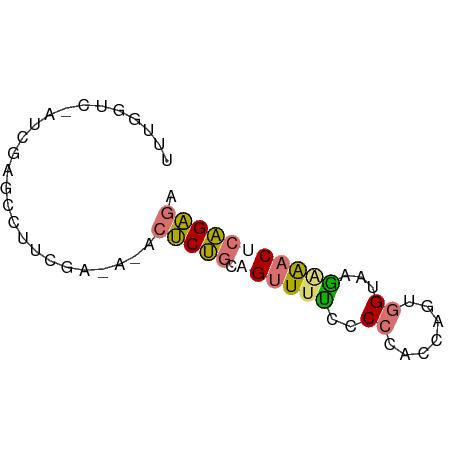
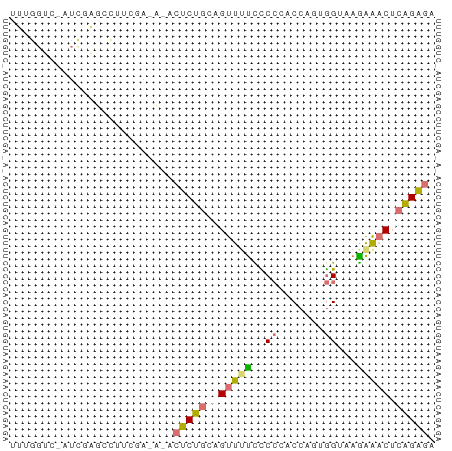
| Location | 14,478,856 – 14,478,921 |
|---|---|
| Length | 65 |
| Sequences | 4 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 52.56 |
| Shannon entropy | 0.79853 |
| G+C content | 0.47333 |
| Mean single sequence MFE | -15.73 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.92 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
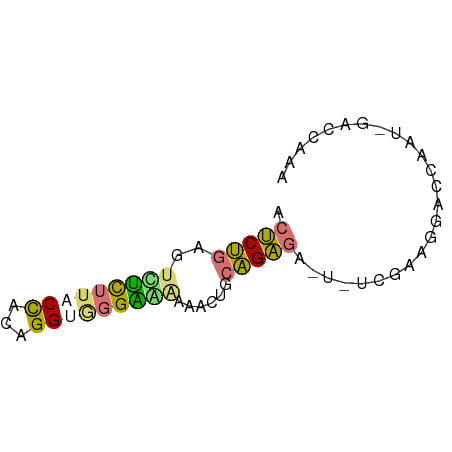
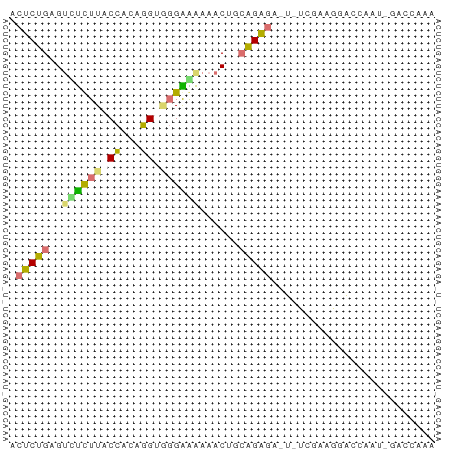
>dm3.chr2R 14478856 65 - 21146708 UCUCUGAGUUCCUUACCUUUGGUAGGGGGAAACUGCAGAGUCGAUUAAAGGCUCGAUUGACCAAA ..(((((((((((((((...))))))))...))).))))(((((((........))))))).... ( -19.70, z-score = -1.28, R) >droSec1.super_43 56432 65 - 327307 UCUCUGAGUUUUUUACUACUGGUGCGAAAAAACUGCAGAGAAUUUCGAAAAACAAAACAACCAAA ((((((((((((((..((....))..)))))))).))))))........................ ( -13.30, z-score = -2.39, R) >droAna3.scaffold_13266 16962352 55 + 19884421 AACCUGAGUCUCUUACCACAGG--GGGGAAAACGUCAGGGU--------GUCUCGAUGGACCAAA ..(((((((.(((..((...))--..)))..)).)))))(.--------((((....)))))... ( -13.70, z-score = 0.78, R) >droPer1.super_164 85937 53 - 94152 ACUCCGUG-CUCUUACCACAGGGGGGAAACAAC---GGAGAGUCUCGAAGGACCAAC-------- .((((((.-(((((.....)))))(....).))---)))).((((....))))....-------- ( -16.20, z-score = -0.90, R) >consensus ACUCUGAGUCUCUUACCACAGGUGGGAAAAAACUGCAGAGA_U_UCGAAGGACCAAU_GACCAAA .(((((..(((((((((...)))))))))......)))))......................... ( -9.99 = -9.92 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:21 2011