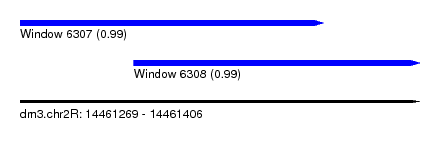
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,461,269 – 14,461,406 |
| Length | 137 |
| Max. P | 0.994627 |

| Location | 14,461,269 – 14,461,373 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 63.79 |
| Shannon entropy | 0.70146 |
| G+C content | 0.37598 |
| Mean single sequence MFE | -22.40 |
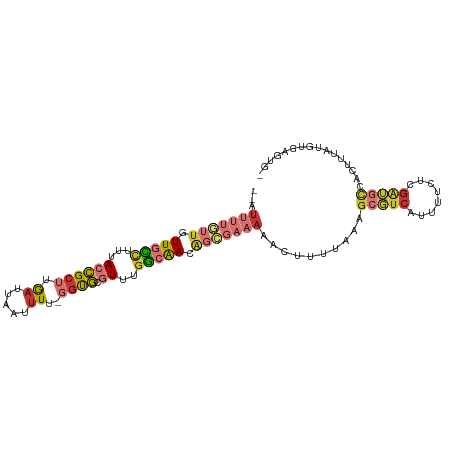
| Consensus MFE | -13.12 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
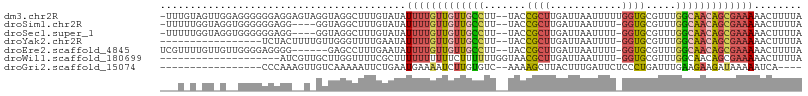
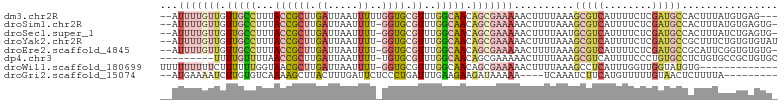
>dm3.chr2R 14461269 104 + 21146708 -UUUGUAGUUGGAGGGGGGAGGAGUAGGUAGGCUUUGUAUAUUUUGUUGUUGCCUU--UACCGCUUGAUUAAUUUUUGGUGCGUUUGGCAACAGCGAAAAACUUUUA -........((((((.....(((((......))))).....(((((((((((((..--.((((((.((......)).)))).))..)))))))))))))..)))))) ( -28.10, z-score = -1.68, R) >droSim1.chr2R 13176938 99 + 19596830 -UUUUUGGUAGGUGGGGGGAGG----GGUAGGCUUUGUAUAUUUUGUUGUUGCCUU--UACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUA -...........((((((((((----......)))).....(((((((((((((..--.((((((.((.....)).-)))).))..)))))))))))))..)))))) ( -25.50, z-score = -1.12, R) >droSec1.super_1 11929886 99 + 14215200 -UUUUUGGUAGGUGGGGGGAGG----GGUAGGCUUUGUAUAUUUUGUUGUUGCCUU--UACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUA -...........((((((((((----......)))).....(((((((((((((..--.((((((.((.....)).-)))).))..)))))))))))))..)))))) ( -25.50, z-score = -1.12, R) >droYak2.chr2R 6411095 87 - 21139217 -----------------UCUACUUUUGUUGGGUUUUGAAUAUUUUGUUGUUGCCUU--UACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUA -----------------((.((((.....))))...))...(((((((((((((..--.((((((.((.....)).-)))).))..)))))))))))))........ ( -24.10, z-score = -3.52, R) >droEre2.scaffold_4845 8663928 98 + 22589142 UCGUUUUGUUGUUGGGGAGGGG------GAGCCUUUGAAUAUUUUGUUGUUGCCUU--UACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUA ..(((((.........(((((.------...))))).......(((((((((((..--.((((((.((.....)).-)))).))..))))))))))))))))..... ( -29.00, z-score = -2.13, R) >droWil1.scaffold_180699 1075864 86 + 2593675 --------------------AUCGUUGCUUGGUUUUCGCUUUUUUUUUUCUUUUUUGGUAACGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUA --------------------............(((((((....................((((((..(......).-.).)))))((....)))))))))....... ( -15.70, z-score = -0.59, R) >droGri2.scaffold_15074 590753 84 + 7742996 -----------------CCCAAAGUUGUCAAAAAUUCUGAAUGAAAAUCUUGUGUC--AAAAGCUUACUUUGAUUCUCCCUGAUUUGAAGAAGAUAAAAAUCA---- -----------------.......(((((.....(((.....)))...........--.........(((..(.((.....)).)..)))..)))))......---- ( -8.90, z-score = 0.38, R) >consensus ___U_U_GU_G__GGGGGGAGG__U_GGUAGGCUUUGUAUAUUUUGUUGUUGCCUU__UACCGCUUGAUUAAUUUU_GGUGCGUUUGGCAACAGCGAAAAACUUUUA .........................................(((((((((((((.......((((.((......)).)))).....)))))))))))))........ (-13.12 = -14.03 + 0.90)
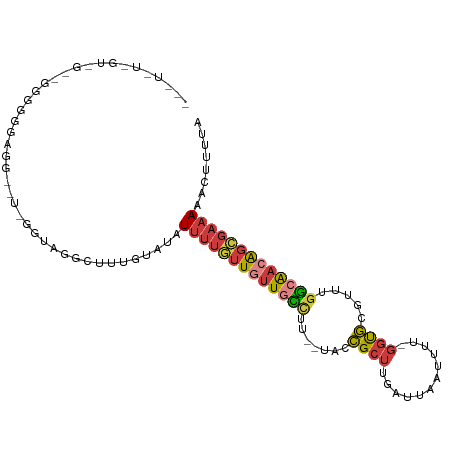
| Location | 14,461,308 – 14,461,406 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.59507 |
| G+C content | 0.37232 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -10.10 |
| Energy contribution | -11.97 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14461308 98 + 21146708 --AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUUUGGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCUCGAUGCCACUUUAUGUGAG--- --.(((((((((((((...((((((.((......)).)))).))..)))))))))))))..........(((((........)))))(((.....)))..--- ( -31.20, z-score = -4.42, R) >droSim1.chr2R 13176973 99 + 19596830 --AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCUCGAUGCCACUUUAUGUGAGUG- --.(((((((((((((...((((((.((.....)).-)))).))..)))))))))))))..........(((((........)))))(((((.....)))))- ( -32.10, z-score = -4.44, R) >droSec1.super_1 11929921 99 + 14215200 --AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUAAUGCGUCAUUUUCUCGAUGCCACUUUAUCUGAGUG- --.(((((((((((((...((((((.((.....)).-)))).))..)))))))))))))..........(((((........)))))(((((.....)))))- ( -31.90, z-score = -4.94, R) >droYak2.chr2R 6411118 100 - 21139217 --AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCUCGAUGCCGCUUUCUGUGUGUAU --.(((((((((((((...((((((.((.....)).-)))).))..))))))))))))).((....((((((.((((.....)))).))))))....)).... ( -32.10, z-score = -4.32, R) >droEre2.scaffold_4845 8663962 99 + 22589142 --AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCUCGAUGCCGCAUUCGGUGUGUG- --.(((((((((((((...((((((.((.....)).-)))).))..)))))))))))))..........(((((........)))))(((((.....)))))- ( -31.80, z-score = -3.26, R) >dp4.chr3 11031057 93 - 19779522 ---------UUUUGUUUUAACCGCUUGAUUAAUUUU-UGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCCCUGUGCCUCUGUGCCGCUGUGC ---------...((((..((((((..((.....)).-.))).))).))))((((((....((.......(((.((.......)))))....))..)))))).. ( -14.80, z-score = 1.20, R) >droWil1.scaffold_180699 1075884 89 + 2593675 UUUUUUUUUCUUUUUUGGUAACGCUUGAUUAAUUUU-GGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCCUCAUUUGGUUGGUAUGUG------------- ....((((((.((.(((.(((((((..(......).-.).))).))).))).)).)))))).......((((......))))........------------- ( -14.50, z-score = 0.46, R) >droGri2.scaffold_15074 590776 88 + 7742996 --AUGAAAAUCUUGUGUCAAAAGCUUACUUUGAUUCUCCCUGAUUUGAAGAAGAUAAAAA----UCAAAUCUUCAUGUUUUUGUAACUCUUUUA--------- --...........((..(((((((....((..(.((.....)).)..))((((((.....----....))))))..)))))))..)).......--------- ( -12.70, z-score = -0.84, R) >consensus __AUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUU_GGUGCGUUUGGCAACAGCGAAAAACUUUUAAAGCGUCAUUUUCUCGAUGCCACUUUAUGUGAGUG_ ...(((((((.(((((...((((((.((......)).)))).))..))))).)))))))..........(((((........)))))................ (-10.10 = -11.97 + 1.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:19 2011