
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,456,645 – 14,456,757 |
| Length | 112 |
| Max. P | 0.949709 |

| Location | 14,456,645 – 14,456,757 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 74.64 |
| Shannon entropy | 0.43590 |
| G+C content | 0.35419 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -14.82 |
| Energy contribution | -16.50 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
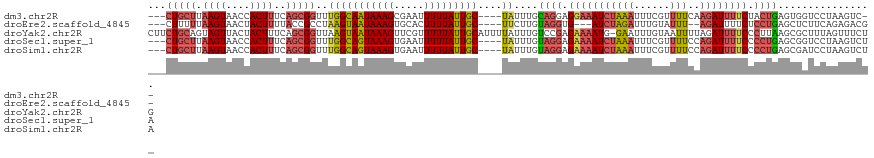
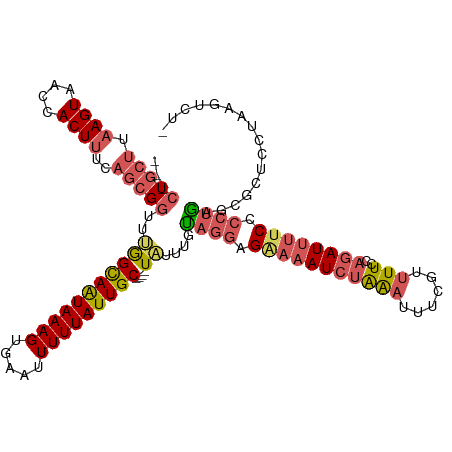
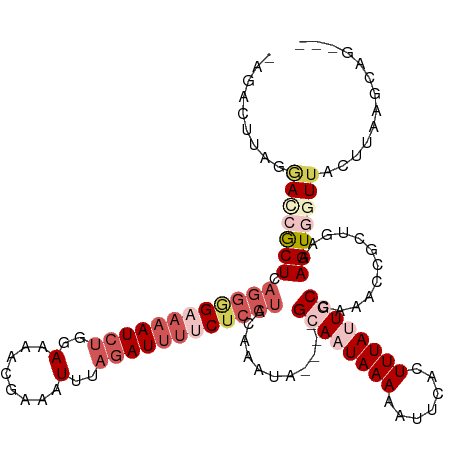
>dm3.chr2R 14456645 112 + 21146708 ---CUGCUUAAGUAACCACUUUCAGCGGUUUGGCAAUAAAGCGAAUUUUUAUUGC----UAUUUGCAGGAGGAAAUCUAAAUUUCGUUUUCAAGAUUUUCUACUGAGUGGUCCUAAGUC-- ---..(((((.(..(((((((...((((..(((((((((((.....)))))))))----)).))))((.((((((((((((......)))..))))))))).)))))))))).))))).-- ( -31.50, z-score = -2.97, R) >droEre2.scaffold_4845 8659354 108 + 22589142 ---CUUUUUAAGUAACUACUUUUACCGCCUAAGUAAUAAAGUGCACUUUUAUUGC----UUCUUGUAGGUG---AUCUAGAUUUGUAUUU--AGAUUUUCUCCUGAGCUCUUCAGAGACG- ---........((((......))))((((((((((((((((.....)))))))))----).....))))))---((((((((....))))--))))..((((.((((...))))))))..- ( -22.60, z-score = -1.37, R) >droYak2.chr2R 6406421 120 - 21139217 CUUCUGCAGUAGUUACUACUUUCAGCGGUUAAGUAAUAAAGUUCGUUUUUAUUGCAUUUUAUUUGUCCGAGAAAAUG-GAAUUUGUAAUUUUAGAUUUUCCCUUAAGCGCUUUAGUUUCUG .........(((..((((.....((((.(((((((((((((.....))))))))).............(((((..((-((((.....))))))..)))))..)))).)))).))))..))) ( -21.40, z-score = -0.65, R) >droSec1.super_1 11925225 114 + 14215200 ---CUGCUUAAGUAACCACUUUCAGCGGUUUGGCAGUAAAGUGAAUUUUUAUUGC----UAUUUGUAGGAGAAAAUCUAAAUUUCGUUUUCCAGAUUUUCCCCUGAGCGGUCCUAAGUCUA ---((((((....((((.(.....).))))(((((((((((.....)))))))))----)).....(((.(((((((((((......)))..)))))))).)))))))))........... ( -29.00, z-score = -2.19, R) >droSim1.chr2R 13172431 114 + 19596830 ---CUGCUUAAGUAACCACUUUCAGCGGUUUGGCAGUAAAGUGAAUUUUUAUUGC----UAUUUGUAGGAGAAAAUCUAAAUUUCGUUUUCCAGAUUUUCCCCUGAGCGAUCCUAAGUCUA ---(((((.((((....))))..)))))..(((((((((((.....)))))))))----)).(((((((.(((((((((((......)))..)))))))).)))..))))........... ( -27.60, z-score = -2.04, R) >consensus ___CUGCUUAAGUAACCACUUUCAGCGGUUUGGCAAUAAAGUGAAUUUUUAUUGC____UAUUUGUAGGAGAAAAUCUAAAUUUCGUUUUCCAGAUUUUCCCCUGAGCGCUCCUAAGUCU_ ...(((((.((((....))))..)))))....(((((((((.....)))))))))..........((((.((((((((..............)))))))).))))................ (-14.82 = -16.50 + 1.68)
| Location | 14,456,645 – 14,456,757 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 74.64 |
| Shannon entropy | 0.43590 |
| G+C content | 0.35419 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -13.32 |
| Energy contribution | -16.72 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
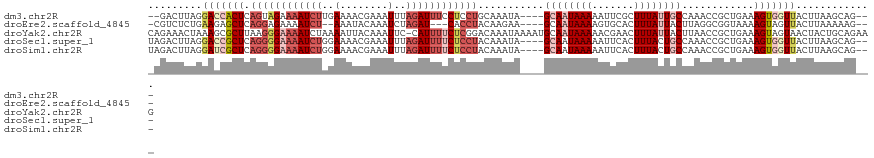
>dm3.chr2R 14456645 112 - 21146708 --GACUUAGGACCACUCAGUAGAAAAUCUUGAAAACGAAAUUUAGAUUUCCUCCUGCAAAUA----GCAAUAAAAAUUCGCUUUAUUGCCAAACCGCUGAAAGUGGUUACUUAAGCAG--- --(.((((((((((((((((((.((((((.((........)).)))))).))..........----((((((((.......))))))))......)))))..)))))..)))))))..--- ( -25.20, z-score = -2.68, R) >droEre2.scaffold_4845 8659354 108 - 22589142 -CGUCUCUGAAGAGCUCAGGAGAAAAUCU--AAAUACAAAUCUAGAU---CACCUACAAGAA----GCAAUAAAAGUGCACUUUAUUACUUAGGCGGUAAAAGUAGUUACUUAAAAAG--- -..(((((((.....))).))))..((((--(.((....)).)))))---...((((.....----(((.......)))..(((((..(....)..))))).))))............--- ( -13.80, z-score = 1.05, R) >droYak2.chr2R 6406421 120 + 21139217 CAGAAACUAAAGCGCUUAAGGGAAAAUCUAAAAUUACAAAUUC-CAUUUUCUCGGACAAAUAAAAUGCAAUAAAAACGAACUUUAUUACUUAACCGCUGAAAGUAGUAACUACUGCAGAAG ..........((((.(((((.....................((-(........)))............((((((.......)))))).))))).))))....(((((....)))))..... ( -16.30, z-score = -0.77, R) >droSec1.super_1 11925225 114 - 14215200 UAGACUUAGGACCGCUCAGGGGAAAAUCUGGAAAACGAAAUUUAGAUUUUCUCCUACAAAUA----GCAAUAAAAAUUCACUUUACUGCCAAACCGCUGAAAGUGGUUACUUAAGCAG--- ..(.((((((((((((.(((((((((((((((........))))))))))))))).......----(((.((((.......)))).)))............))))))..)))))))..--- ( -32.20, z-score = -4.09, R) >droSim1.chr2R 13172431 114 - 19596830 UAGACUUAGGAUCGCUCAGGGGAAAAUCUGGAAAACGAAAUUUAGAUUUUCUCCUACAAAUA----GCAAUAAAAAUUCACUUUACUGCCAAACCGCUGAAAGUGGUUACUUAAGCAG--- ..(.((((((.......(((((((((((((((........))))))))))))))).......----(((.((((.......)))).)))..((((((.....)))))).)))))))..--- ( -32.20, z-score = -4.14, R) >consensus _AGACUUAGGACCGCUCAGGGGAAAAUCUGGAAAACGAAAUUUAGAUUUUCUCCUACAAAUA____GCAAUAAAAAUUCACUUUAUUGCCAAACCGCUGAAAGUGGUUACUUAAGCAG___ .........(((((((.((((((((((((.((........)).))))))))))))...........((((((((.......))))))))............)))))))............. (-13.32 = -16.72 + 3.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:18 2011