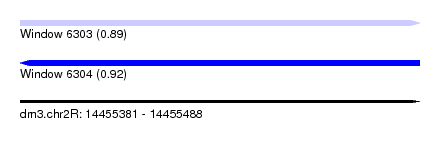
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,455,381 – 14,455,488 |
| Length | 107 |
| Max. P | 0.918078 |

| Location | 14,455,381 – 14,455,488 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.55 |
| Shannon entropy | 0.16694 |
| G+C content | 0.59100 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -33.14 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
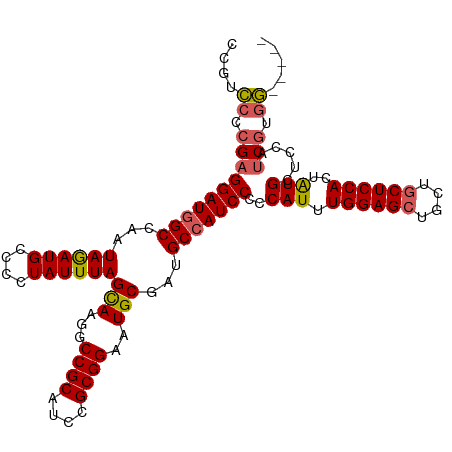
>dm3.chr2R 14455381 107 + 21146708 CCGUCCCCGAGGAUUGCCAAUAGAUGCCCCUAUUUAGCAAGGCCGCAUCCGCGGAAUGCGAUGCUAUCCCCCAUUUGGAGCUGCUGCUCCACUAUGCUCCAUCGUGG----- .....(((((((((.((.(((((......)))))..(((...((((....))))..)))...)).))))..(((.((((((....))))))..))).....))).))----- ( -32.90, z-score = -1.01, R) >droEre2.scaffold_4845 8658110 107 + 22589142 CCGUCCCCGAGGAUGGCCAAUAGAUGCCCCUAUUUAGCAAGGCCGCAGCCGCGGAAUGCGAUGCCAUCCCCCAUUUGGAGCUGCUGCUCCACAAUGCUCCGACGUGG----- .((((.....(((((((.(((((......)))))..(((...((((....))))..)))...)))))))..((((((((((....)))))).))))....))))...----- ( -40.70, z-score = -2.36, R) >droYak2.chr2R 6405090 112 - 21139217 UCGUGCCCGAGGAUGGCCAAUAGAUGCCCCUAUUUAGUAAGGCCGCAUCCGCGGAAUGCGCUGCCAUCCCCCAUUUGGAGCUGCUGCUCCACUAUGUACUAUCCAUCCAGUG ..((((....(((((((.....(((((.(((........)))..))))).(((.....))).)))))))......((((((....))))))....))))............. ( -37.00, z-score = -1.93, R) >droSec1.super_1 11923956 107 + 14215200 CCGUCCCCGAGGAUGGCCAAUAAAUGCCCCUAUUUAGCAAGGCCGCAUCCGCGGAAUGCGAUGCCAUCCCCCAUUUGGAGAUGCUGCUCCACUGUGCUCCAUCGCGG----- ((((..(((((((((((...((((((....))))))(((...((((....))))..)))...))))))).....)))).((((..((........))..))))))))----- ( -36.10, z-score = -1.31, R) >droSim1.chr2R 13171162 107 + 19596830 CCGUCCCCGAGGAUGGCCAAUAGAUGCCCCUAUUUAGCAAGGCCGCAUCCGCGGAAUGCGAUGCCAUCCCCCAUUUGGAGCUGCUGCUCCACUAUGCUCCAUCGUGG----- .....((((((((((((.(((((......)))))..(((...((((....))))..)))...)))))))..(((.((((((....))))))..))).....))).))----- ( -39.40, z-score = -2.23, R) >consensus CCGUCCCCGAGGAUGGCCAAUAGAUGCCCCUAUUUAGCAAGGCCGCAUCCGCGGAAUGCGAUGCCAUCCCCCAUUUGGAGCUGCUGCUCCACUAUGCUCCAUCGUGG_____ ....((.((((((((((...((((((....))))))(((...((((....))))..)))...)))))))..(((.((((((....))))))..))).....))).))..... (-33.14 = -33.34 + 0.20)

| Location | 14,455,381 – 14,455,488 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Shannon entropy | 0.16694 |
| G+C content | 0.59100 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -40.28 |
| Energy contribution | -40.88 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
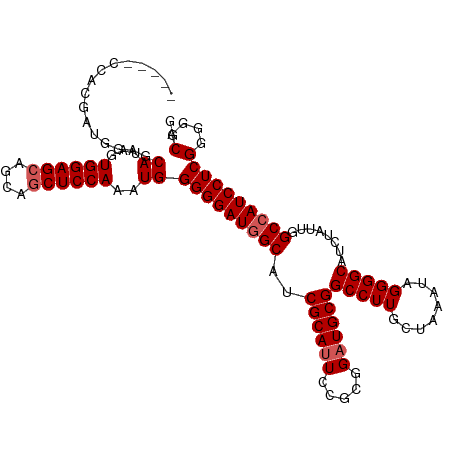
>dm3.chr2R 14455381 107 - 21146708 -----CCACGAUGGAGCAUAGUGGAGCAGCAGCUCCAAAUGGGGGAUAGCAUCGCAUUCCGCGGAUGCGGCCUUGCUAAAUAGGGGCAUCUAUUGGCAAUCCUCGGGGACGG -----((.(((.(((.(((..((((((....)))))).)))((((...(((((((.....)).)))))..))))(((((.((((....)))))))))..))))))))..... ( -39.20, z-score = -1.07, R) >droEre2.scaffold_4845 8658110 107 - 22589142 -----CCACGUCGGAGCAUUGUGGAGCAGCAGCUCCAAAUGGGGGAUGGCAUCGCAUUCCGCGGCUGCGGCCUUGCUAAAUAGGGGCAUCUAUUGGCCAUCCUCGGGGACGG -----...((((....((((.((((((....))))))))))(((((((((...(((..((((....))))...)))..((((((....)))))).)))))))))...)))). ( -47.80, z-score = -2.20, R) >droYak2.chr2R 6405090 112 + 21139217 CACUGGAUGGAUAGUACAUAGUGGAGCAGCAGCUCCAAAUGGGGGAUGGCAGCGCAUUCCGCGGAUGCGGCCUUACUAAAUAGGGGCAUCUAUUGGCCAUCCUCGGGCACGA .((((......)))).(((..((((((....)))))).)))(((((((((..((((((.....))))))(((((........)))))........)))))))))........ ( -44.20, z-score = -2.11, R) >droSec1.super_1 11923956 107 - 14215200 -----CCGCGAUGGAGCACAGUGGAGCAGCAUCUCCAAAUGGGGGAUGGCAUCGCAUUCCGCGGAUGCGGCCUUGCUAAAUAGGGGCAUUUAUUGGCCAUCCUCGGGGACGG -----((((((((..((........))..(((((((.....))))))).)))))).(((((.((((..((((.((((.......))))......)))))))).)))))..)) ( -42.10, z-score = -1.19, R) >droSim1.chr2R 13171162 107 - 19596830 -----CCACGAUGGAGCAUAGUGGAGCAGCAGCUCCAAAUGGGGGAUGGCAUCGCAUUCCGCGGAUGCGGCCUUGCUAAAUAGGGGCAUCUAUUGGCCAUCCUCGGGGACGG -----((.....))..(((..((((((....)))))).)))(((((((((..((((((.....))))))(((((........)))))........)))))))))(....).. ( -45.20, z-score = -2.05, R) >consensus _____CCACGAUGGAGCAUAGUGGAGCAGCAGCUCCAAAUGGGGGAUGGCAUCGCAUUCCGCGGAUGCGGCCUUGCUAAAUAGGGGCAUCUAUUGGCCAUCCUCGGGGACGG ................((...((((((....))))))..))(((((((((..((((((.....))))))(((((........)))))........)))))))))(....).. (-40.28 = -40.88 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:16 2011