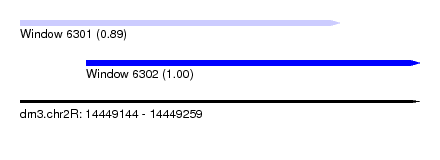
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,449,144 – 14,449,259 |
| Length | 115 |
| Max. P | 0.996150 |

| Location | 14,449,144 – 14,449,236 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.57829 |
| G+C content | 0.54759 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -13.17 |
| Energy contribution | -15.24 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14449144 92 + 21146708 AACGAGACCCAGUCAGAACGUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG--UCCGAAACGAAGUUGGAGCUAAAGCUUCG-CCUCGGAG ...((...(((((....)).)))...))((((((((((....))))))))))...--(((((..(((((((........)))))))-..))))). ( -32.60, z-score = -1.97, R) >droSim1.chr2R 13164463 94 + 19596830 AACGAGCCCCAGUCAGAACGUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCGCGUCCGAAUCGAAGUUGGAGCUGAAGCUUCG-CCUCGGAG ..................(((((.....((((((((((....)))))))))))))))(((((..(((((((........)))))))-..))))). ( -35.50, z-score = -1.97, R) >droYak2.chr2R 6397977 89 - 21139217 AACGAGCUGCAGUCAGAACGUGGCCAUCAGUCAGUGUGUGUGCAUUCUGGCUCCG------AGUCCAAGUUGGAGCUGAAGCCUUGUGCUCGGAG ...(.((..(.((....)))..)))....(((((.(((....))).)))))((((------((..((((.....((....))))))..)))))). ( -28.80, z-score = -0.12, R) >droEre2.scaffold_4845 8651697 88 + 22589142 AACGAGCCGCAGUCAGAACGUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG------AGUCGAAGCUGGAGCUGAAGCUUUG-GCUCGGAG ...(.(((((.((....))))))))....(((((((((....)))))))))((((------((((((((((........)))))))-))))))). ( -44.20, z-score = -4.29, R) >droGri2.scaffold_15245 10492409 70 + 18325388 -----AACAAGAAUAAAACGUGGCCAUCAGUCAAGGCACAAAAGGCAAGCCUUCA------ACUUCGGCAUCGACUUCAGA-------------- -----.............((..(((...(((.(((((...........)))))..------)))..)))..))........-------------- ( -12.00, z-score = -0.42, R) >consensus AACGAGCCCCAGUCAGAACGUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG______AAUCGAAGUUGGAGCUGAAGCUUCG_CCUCGGAG ..((((...............((.....((((((((((....))))))))))))..........(((((((........)))))))..))))... (-13.17 = -15.24 + 2.07)
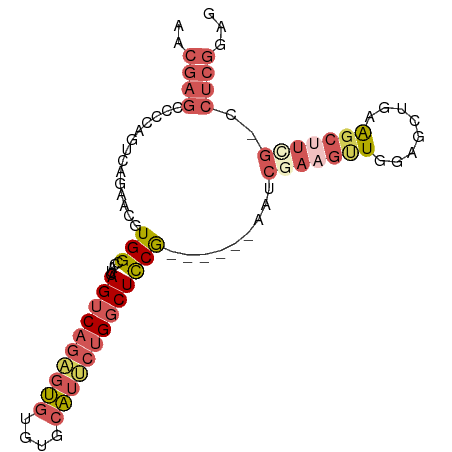
| Location | 14,449,163 – 14,449,259 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.58296 |
| G+C content | 0.55008 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -20.12 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14449163 96 + 21146708 GUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG--UCCGAAACGAAGUUGGAGCUAAAGCUUCGCCUCGGAGCUGGACCUGGAACUGUAGCUGGA ..(((((.(((((((((((....))))))))))...--(((((..(((((((........)))))))..)))))).))).))................ ( -35.00, z-score = -1.07, R) >droSim1.chr2R 13164482 98 + 19596830 GUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCGCGUCCGAAUCGAAGUUGGAGCUGAAGCUUCGCCUCGGAGCUGGAGCUGGAACUGUUGCUGGA ..(((...(((((((((((....)))))))(((((((.(((((..(((((((........)))))))..))))))).)))))....))))..)))... ( -42.80, z-score = -2.35, R) >droEre2.scaffold_4845 8651716 87 + 22589142 GUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG------AGUCGAAGCUGGAGCUGAAGCUUUGGCUCGGAGCU-----UGGAACUGGUGCUGGA ..(((.(((((((((((((....)))))))((((((------((((((((((........)))))))))))))))).-----....)))))))))... ( -47.80, z-score = -5.31, R) >droMoj3.scaffold_6496 23412238 81 - 26866924 GCGGCCAUCAGUCAAGGCA-ACAAGUCAAGACAUCGACUUCUACUUCUGCUUGGAGUUCGAGUUUGAGCUCG-AGUUUGAGUU--------------- (..(((.........))).-.)(((((........)))))........(((..((.(((((((....)))))-))))..))).--------------- ( -21.20, z-score = -0.01, R) >consensus GUGGCCAUCAGUCAGAGUGUGUGCAUUCUGGCUCCG__UCCGAAUCGAAGUUGGAGCUGAAGCUUCGCCUCGGAGCUGGAGCUGGAACUGUUGCUGGA ..(((.....(((((((((....)))))))))......(((((.((((((((........)))))))).))))))))..................... (-20.12 = -21.00 + 0.87)

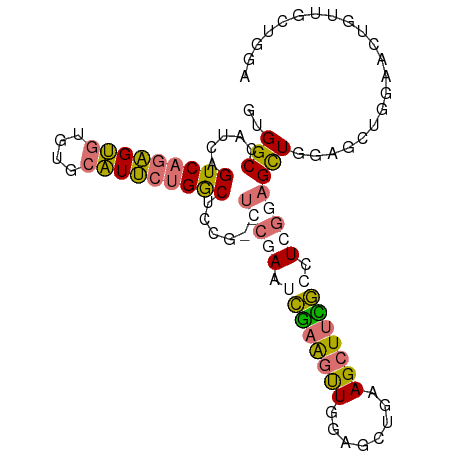
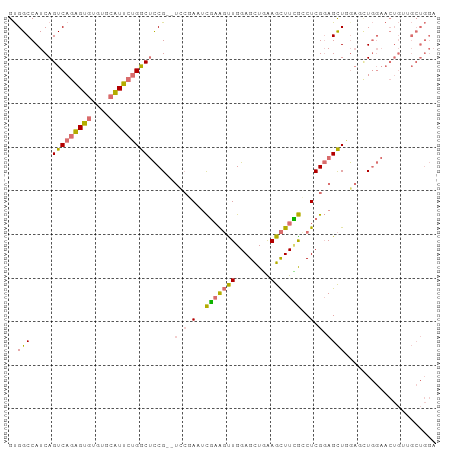
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:14 2011