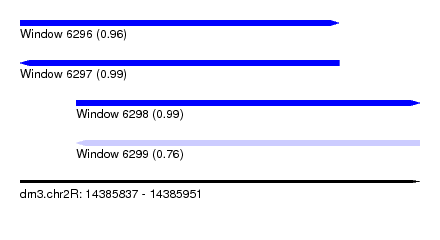
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,385,837 – 14,385,951 |
| Length | 114 |
| Max. P | 0.989543 |

| Location | 14,385,837 – 14,385,928 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.54 |
| Shannon entropy | 0.59721 |
| G+C content | 0.47223 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -12.91 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
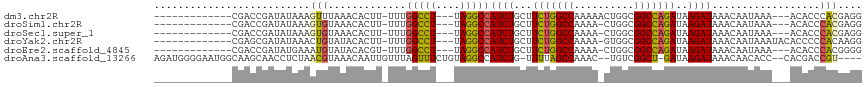
>dm3.chr2R 14385837 91 + 21146708 ---------------------UCGAGGCAAUUU-AUAUCGACCGAUAUAAAGUUUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAAACUGGCGGCCAGAUAAGA ---------------------....(((..(((-(((((....)))))))).......((.((-(((((((.---.((((.....))))..))))))))).))...)))........ ( -30.00, z-score = -3.45, R) >droSim1.chr2R 13103243 89 + 19596830 ---------------------UCGAC-CAAUUA-AUAUCGACCGAUAUAAAGUGUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGA ---------------------.....-......-(((((....)))))((((((....)))))-).(((((.---..)))))(((...(((((((....-.....)))))))..))) ( -27.90, z-score = -3.41, R) >droSec1.super_1 11854877 89 + 14215200 ---------------------UCGAC-CAAUUA-AUAUCGACCGAUAUAAAGUGUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGA ---------------------.....-......-(((((....)))))((((((....)))))-).(((((.---..)))))(((...(((((((....-.....)))))))..))) ( -27.90, z-score = -3.41, R) >droYak2.chr2R 6331220 89 - 21139217 ----------------------UAGGCCAAUUA-AUAUCGAGCGAUAUAAACUGUAUACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-GUGGCGGCCAGAUAAGA ----------------------..((((.....-(((((....)))))..........(((((-((.((((.---.((((.....))))..))))))))-)))..))))........ ( -31.70, z-score = -3.39, R) >droEre2.scaffold_4845 8586139 89 + 22589142 ---------------------UCGGC-CAAUUA-AUAUCGACCGAUAUGAAAUGUAUACACGU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGA ---------------------..(((-(.....-(((((....)))))..........((.((-(((((((.---.((((.....))))..))))))))-)))..))))........ ( -31.80, z-score = -3.68, R) >droAna3.scaffold_13266 13213725 111 - 19884421 UUGUGGCAAUUAAUACCA-AAAAGAGAUGGGGA-AUGGCAAGCAACCUCUAACGUAAACAAUUGUUUAGUUUCUGUAGGCCAUCUG-UUUUAGCCAAA--CUGUCGGCU-GAUAAGA ...((((......(((..-...((((((((((.-...(....)..)))).....(((((....)))))))))))))).))))(((.-..((((((...--.....))))-))..))) ( -23.00, z-score = 0.55, R) >dp4.chr3 11627944 107 + 19779522 -GGGGGCCAUUAGCGUCAGAAACGCCACAGCCGCAGAAUGAUUGGGCGCGAGUGUAAACACUU-UUCGACCU---CCGGCCAUCUG-UGCGGGCCAA---CUGGCGGCU-GAUAAGA -.......(((((((((((....(((...((.(((((.((..((((.(((((((....)))..-.))).).)---)))..))))))-))).)))...---))))).)))-))).... ( -35.20, z-score = 0.49, R) >droPer1.super_4 7026814 107 + 7162766 -GGGGGCCAUCAGCGUCAGAAACGCCACAACCGCAGAAUGAUUGGGCGCGAGUGUAAACACUU-UUCGACCU---CCGGCCAUCUG-UGCGGGCCAA---CUGGCGGCU-GAUAAGA -.......(((((((((((....(((.....((((((.((..((((.(((((((....)))..-.))).).)---)))..))))))-))..)))...---))))).)))-))).... ( -35.70, z-score = 0.15, R) >consensus _____________________UCGAC_CAAUUA_AUAUCGACCGAUAUAAAGUGUAAACACUU_UUUGGCCU___UAGGCCAUCUGCUUCUGGCCAAAA_CUGGCGGCCAGAUAAGA ...................................................................((((......)))).(((...((.((((..........)))).))..))) (-12.91 = -12.86 + -0.05)

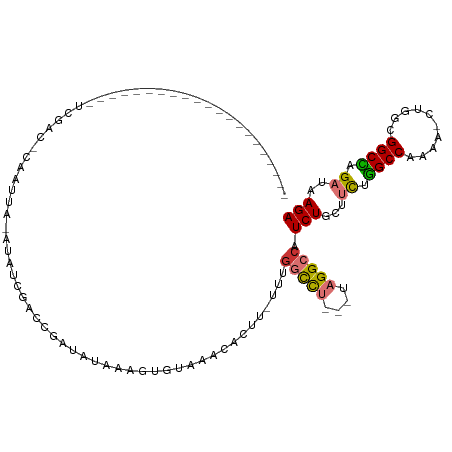
| Location | 14,385,837 – 14,385,928 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.54 |
| Shannon entropy | 0.59721 |
| G+C content | 0.47223 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -13.64 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14385837 91 - 21146708 UCUUAUCUGGCCGCCAGUUUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUAAACUUUAUAUCGGUCGAUAU-AAAUUGCCUCGA--------------------- .....((((((((........)))))))).((((.(((((..---.)))))..-............((((((((....)))))-))))))).....--------------------- ( -28.90, z-score = -2.74, R) >droSim1.chr2R 13103243 89 - 19596830 UCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUACACUUUAUAUCGGUCGAUAU-UAAUUG-GUCGA--------------------- (((..((((((((....-...))))))))...)))(((((..---.))))).(-(((((....))))))...(((..((((..-..))))-..)))--------------------- ( -32.60, z-score = -3.98, R) >droSec1.super_1 11854877 89 - 14215200 UCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUACACUUUAUAUCGGUCGAUAU-UAAUUG-GUCGA--------------------- (((..((((((((....-...))))))))...)))(((((..---.))))).(-(((((....))))))...(((..((((..-..))))-..)))--------------------- ( -32.60, z-score = -3.98, R) >droYak2.chr2R 6331220 89 + 21139217 UCUUAUCUGGCCGCCAC-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUAUACAGUUUAUAUCGCUCGAUAU-UAAUUGGCCUA---------------------- .....((((((((....-...)))))))).((...(((((..---.)))))..-.........(((((.(((((....)))))-.)))))))...---------------------- ( -28.70, z-score = -2.53, R) >droEre2.scaffold_4845 8586139 89 - 22589142 UCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-ACGUGUAUACAUUUCAUAUCGGUCGAUAU-UAAUUG-GCCGA--------------------- (((..((((((((....-...))))))))...)))(((((..---.)))))..-..................(((((((((..-..))))-)))))--------------------- ( -30.80, z-score = -3.01, R) >droAna3.scaffold_13266 13213725 111 + 19884421 UCUUAUC-AGCCGACAG--UUUGGCUAAAA-CAGAUGGCCUACAGAAACUAAACAAUUGUUUACGUUAGAGGUUGCUUGCCAU-UCCCCAUCUCUUUU-UGGUAUUAAUUGCCACAA .......-((((((...--.))))))((((-.((((((........((((((((....))))).)))...(((.....)))..-...)))))).))))-(((((.....)))))... ( -25.40, z-score = -1.49, R) >dp4.chr3 11627944 107 - 19779522 UCUUAUC-AGCCGCCAG---UUGGCCCGCA-CAGAUGGCCGG---AGGUCGAA-AAGUGUUUACACUCGCGCCCAAUCAUUCUGCGGCUGUGGCGUUUCUGACGCUAAUGGCCCCC- ......(-((((((.((---((((((....-.....))))))---.((.((..-.((((....))))..)).)).......)))))))))(((((((...))))))).........- ( -35.40, z-score = -0.28, R) >droPer1.super_4 7026814 107 - 7162766 UCUUAUC-AGCCGCCAG---UUGGCCCGCA-CAGAUGGCCGG---AGGUCGAA-AAGUGUUUACACUCGCGCCCAAUCAUUCUGCGGUUGUGGCGUUUCUGACGCUGAUGGCCCCC- ..(((((-(((.((((.---(((.......-))).))))...---..((((((-.((((....)))).((((((((((.......))))).)))))))).))))))))))).....- ( -35.80, z-score = -0.28, R) >consensus UCUUAUCUGGCCGCCAG_UUUUGGCCAGAAGCAGAUGGCCUA___AGGCCAAA_AAGUGUUUACACUUUAUAUCGGUCGAUAU_UAAUUG_GUCGA_____________________ (((..((.(((((........))))).))...)))(((((......)))))....((((....)))).................................................. (-13.64 = -14.19 + 0.55)
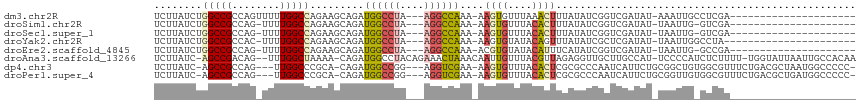
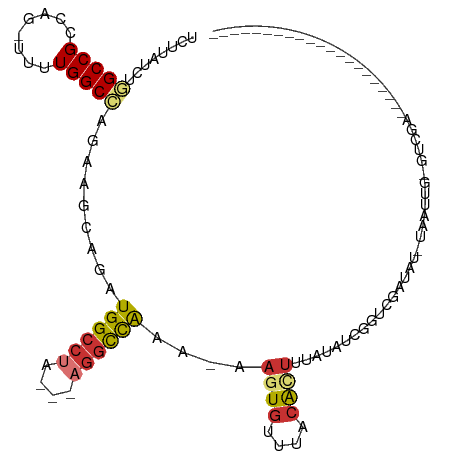
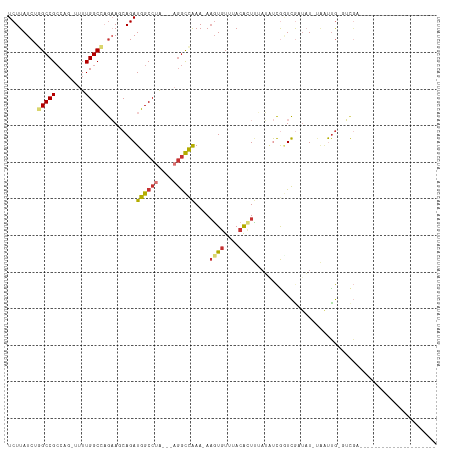
| Location | 14,385,853 – 14,385,951 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.34765 |
| G+C content | 0.45090 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14385853 98 + 21146708 -------------CGACCGAUAUAAAGUUUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAAACUGGCGGCCAGAUAAGAUAAACAAUAAA---ACACCCACGAGG -------------.............(((((.......-..(((((.---..)))))(((...(((((((..........)))))))..))))))))......---............ ( -24.50, z-score = -2.36, R) >droSim1.chr2R 13103258 97 + 19596830 -------------CGACCGAUAUAAAGUGUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGAUAAACAAUAAA---ACACCCACGAGG -------------...((.....((((((....)))))-)..((((.---..))))((((...(((((((....-.....)))))))..))))..........---..........)) ( -26.90, z-score = -2.80, R) >droSec1.super_1 11854892 97 + 14215200 -------------CGACCGAUAUAAAGUGUAAACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGAUAAACAAUAAA---ACACCCACGAGG -------------...((.....((((((....)))))-)..((((.---..))))((((...(((((((....-.....)))))))..))))..........---..........)) ( -26.90, z-score = -2.80, R) >droYak2.chr2R 6331235 100 - 21139217 -------------CGAGCGAUAUAAACUGUAUACACUU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-GUGGCGGCCAGAUAAGAUAAACAAUAAAUACACCCCCACAAGG -------------..............(((((......-...((((.---..))))((((...(((((((....-.....)))))))..)))).........)))))........... ( -24.60, z-score = -1.38, R) >droEre2.scaffold_4845 8586154 97 + 22589142 -------------CGACCGAUAUGAAAUGUAUACACGU-UUUGGCCU---UAGGCCAUCUGCUUCUGGCCAAAA-CUGGCGGCCAGAUAAGAUAAACAAUAAA---ACACCCACGGGG -------------...(((....(((((((....))))-)))((((.---..))))((((...(((((((....-.....)))))))..))))..........---.......))).. ( -29.20, z-score = -2.74, R) >droAna3.scaffold_13266 13213748 108 - 19884421 AGAUGGGGAAUGGCAAGCAACCUCUAACGUAAACAAUUGUUUAGUUUCUGUAGGCCAUCUG-UUUUAGCCAAAC--UGUCGGCU-GAUAAGAUAAACAACACC--CACGACCGU---- ...((((....(((..(((......(((.(((((....))))))))..)))..)))((((.-..((((((....--....))))-))..))))........))--)).......---- ( -27.30, z-score = -1.04, R) >consensus _____________CGACCGAUAUAAAGUGUAAACACUU_UUUGGCCU___UAGGCCAUCUGCUUCUGGCCAAAA_CUGGCGGCCAGAUAAGAUAAACAAUAAA___ACACCCACGAGG ..........................(((.............(((((....)))))((((...(((((((..........)))))))..))))..................))).... (-21.52 = -21.22 + -0.30)
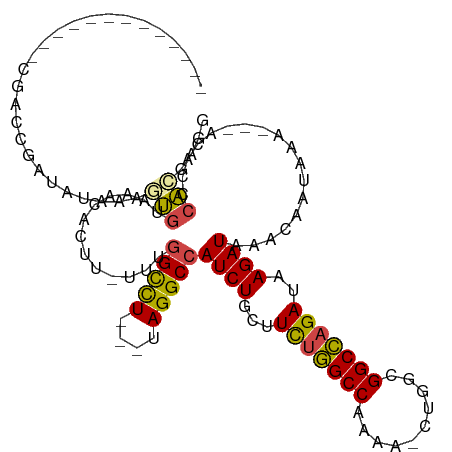
| Location | 14,385,853 – 14,385,951 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.34765 |
| G+C content | 0.45090 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14385853 98 - 21146708 CCUCGUGGGUGU---UUUAUUGUUUAUCUUAUCUGGCCGCCAGUUUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUAAACUUUAUAUCGGUCG------------- ......((.((.---..((..(((((.(((.((((((((........))))))))......(((((..---.)))))..-)))....)))))..))...)).)).------------- ( -26.70, z-score = -1.39, R) >droSim1.chr2R 13103258 97 - 19596830 CCUCGUGGGUGU---UUUAUUGUUUAUCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUACACUUUAUAUCGGUCG------------- .....(((((..---.........(((((..((((((((....-...))))))))...))))))))))---.((((..(-(((((....)))))).....)))).------------- ( -27.90, z-score = -1.62, R) >droSec1.super_1 11854892 97 - 14215200 CCUCGUGGGUGU---UUUAUUGUUUAUCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUUUACACUUUAUAUCGGUCG------------- .....(((((..---.........(((((..((((((((....-...))))))))...))))))))))---.((((..(-(((((....)))))).....)))).------------- ( -27.90, z-score = -1.62, R) >droYak2.chr2R 6331235 100 + 21139217 CCUUGUGGGGGUGUAUUUAUUGUUUAUCUUAUCUGGCCGCCAC-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-AAGUGUAUACAGUUUAUAUCGCUCG------------- ((....))((((((((..(((((.((((((.((((((((....-...))))))))......(((((..---.)))))..-))).))).)))))..))).))))).------------- ( -33.90, z-score = -2.63, R) >droEre2.scaffold_4845 8586154 97 - 22589142 CCCCGUGGGUGU---UUUAUUGUUUAUCUUAUCUGGCCGCCAG-UUUUGGCCAGAAGCAGAUGGCCUA---AGGCCAAA-ACGUGUAUACAUUUCAUAUCGGUCG------------- ..(((.((((((---.....(((((((((..((((((((....-...))))))))...))))((((..---.)))).))-))).....)))))).....)))...------------- ( -28.70, z-score = -1.68, R) >droAna3.scaffold_13266 13213748 108 + 19884421 ----ACGGUCGUG--GGUGUUGUUUAUCUUAUC-AGCCGACA--GUUUGGCUAAAA-CAGAUGGCCUACAGAAACUAAACAAUUGUUUACGUUAGAGGUUGCUUGCCAUUCCCCAUCU ----..(((((((--(((.((((((........-((((((..--..)))))).)))-)))...))))))...((((((((....))))).))).))(((.....))).....)).... ( -24.20, z-score = 0.31, R) >consensus CCUCGUGGGUGU___UUUAUUGUUUAUCUUAUCUGGCCGCCAG_UUUUGGCCAGAAGCAGAUGGCCUA___AGGCCAAA_AAGUGUUUACACUUUAUAUCGGUCG_____________ ....(((..................((((..((((((((........))))))))...))))(((((....))))).............))).......................... (-20.90 = -20.93 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:12 2011