| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,328,366 – 14,328,441 |
| Length | 75 |
| Max. P | 0.861627 |

| Location | 14,328,366 – 14,328,441 |
|---|---|
| Length | 75 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 61.49 |
| Shannon entropy | 0.60628 |
| G+C content | 0.37417 |
| Mean single sequence MFE | -13.85 |
| Consensus MFE | -6.15 |
| Energy contribution | -5.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
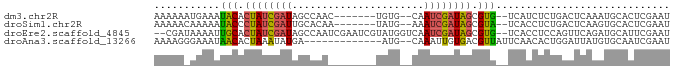
>dm3.chr2R 14328366 75 + 21146708 AAAAAAUGAAAUACACUAUCGAUAGCCAAC-------UGUG--CAAUCGAUAGCGUG--UCAUCUCUGACUCAAAUGCACUCGAAU ......(((...((.((((((((.(((...-------.).)--).)))))))).))(--(((....)))))))............. ( -17.30, z-score = -2.93, R) >droSim1.chr2R 13034692 75 + 19596830 AAAAACAAAAAUACCCUAUCGAUUGCACAA-------UAUG--AAAUCGAUAGCGUA--UCACCUCUGACUCAAGUGCACUCGAAU ..........((((.(((((((((.((...-------..))--.))))))))).)))--)..........((.((....)).)).. ( -13.30, z-score = -2.87, R) >droEre2.scaffold_4845 8535926 82 + 22589142 --CGAUAAAAUUGCACUAUCGAUAGCCAAUCGAAUCGUAUGGUCAAUCGAUAGCGUG--UCACCUCCAGUUCAGAUGCAUUCGAAU --(((..........((((((((.((((..((...))..))))..)))))))).(((--((............)))))..)))... ( -17.00, z-score = -0.53, R) >droAna3.scaffold_13266 15682125 71 - 19884421 AAAAGGGAAAUAACACUAAAUAUGA-------------AUG--CAAAUUGUGACGUUAUUCAACACUGGAUUAUGUGCAAUCGAAU .........................-------------...--...(((((.((((.(((((....))))).)))))))))..... ( -7.80, z-score = 0.34, R) >consensus AAAAAAAAAAAUACACUAUCGAUAGCCAA________UAUG__CAAUCGAUAGCGUG__UCACCUCUGACUCAAAUGCACUCGAAU ...........(((.((((((((......................)))))))).)))............................. ( -6.15 = -5.90 + -0.25)

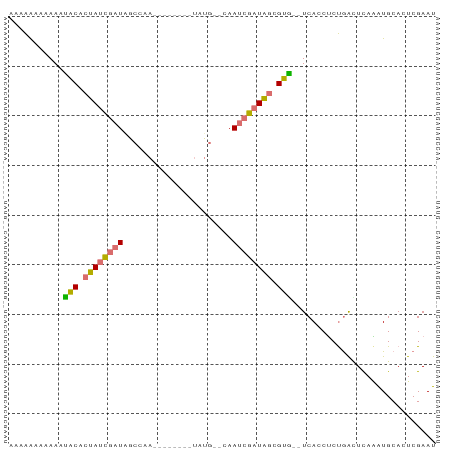
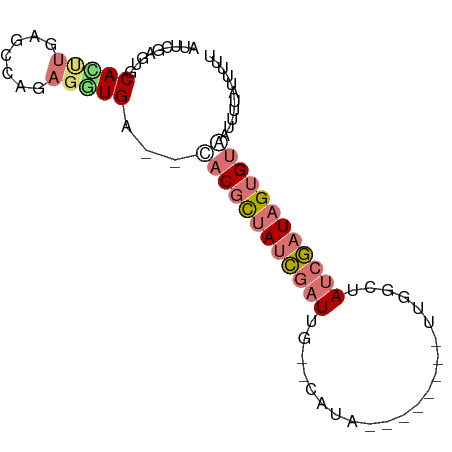
| Location | 14,328,366 – 14,328,441 |
|---|---|
| Length | 75 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 61.49 |
| Shannon entropy | 0.60628 |
| G+C content | 0.37417 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -6.39 |
| Energy contribution | -7.70 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14328366 75 - 21146708 AUUCGAGUGCAUUUGAGUCAGAGAUGA--CACGCUAUCGAUUG--CACA-------GUUGGCUAUCGAUAGUGUAUUUCAUUUUUU (((((((....))))))).((((((((--.(((((((((((.(--(.(.-------...))).)))))))))))...)))))))). ( -24.10, z-score = -3.29, R) >droSim1.chr2R 13034692 75 - 19596830 AUUCGAGUGCACUUGAGUCAGAGGUGA--UACGCUAUCGAUUU--CAUA-------UUGUGCAAUCGAUAGGGUAUUUUUGUUUUU (((((((....)))))))((((...((--(((.(((((((((.--(((.-------..))).))))))))).)))))))))..... ( -23.10, z-score = -3.44, R) >droEre2.scaffold_4845 8535926 82 - 22589142 AUUCGAAUGCAUCUGAACUGGAGGUGA--CACGCUAUCGAUUGACCAUACGAUUCGAUUGGCUAUCGAUAGUGCAAUUUUAUCG-- ...(((.(((((((.......))))).--))((((((((((.(.(((..((...))..)))).))))))))))........)))-- ( -21.00, z-score = -1.29, R) >droAna3.scaffold_13266 15682125 71 + 19884421 AUUCGAUUGCACAUAAUCCAGUGUUGAAUAACGUCACAAUUUG--CAU-------------UCAUAUUUAGUGUUAUUUCCCUUUU ....((.(((((((......))))(((......)))......)--)).-------------))....................... ( -6.10, z-score = 0.39, R) >consensus AUUCGAGUGCACUUGAGCCAGAGGUGA__CACGCUAUCGAUUG__CAUA________UUGGCUAUCGAUAGUGUAAUUUUAUUUUU .........(((((.......)))))....(((((((((((......................)))))))))))............ ( -6.39 = -7.70 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:07 2011