
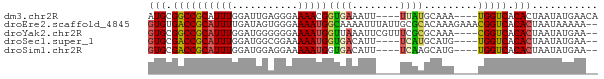
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,327,989 – 14,328,056 |
| Length | 67 |
| Max. P | 0.703308 |

| Location | 14,327,989 – 14,328,056 |
|---|---|
| Length | 67 |
| Sequences | 5 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Shannon entropy | 0.38956 |
| G+C content | 0.42561 |
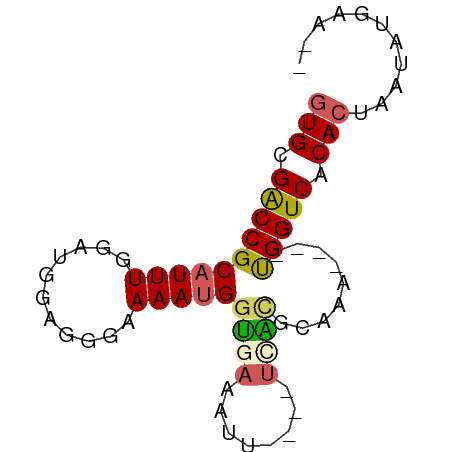
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14327989 67 + 21146708 AUGCGGCCGCAUUUGGAUUGAGGGAAAACGGUGAAAUU----UUAUGCAAA----UGGUCACACUAAUAUGAACA .((.((((((((..(((((..(......).....))))----).))))...----.)))).))............ ( -9.00, z-score = 1.15, R) >droEre2.scaffold_4845 8535845 73 + 22589142 GUGUGACCGCAUUUUGAUAGUGGGAAAAUGGCAAAAUUUAUUGCGCACAAAGAAACGGUCACACUAAUAAAAA-- (((((((((..(((((...(((.((((((......)))).)).))).)))))...))))))))).........-- ( -21.10, z-score = -3.63, R) >droYak2.chr2R 6280766 69 - 21139217 GUGCGGCCGCAUUUGGAUGGGGGGAAAAUGGUUAAAUUCGUUUCGCGCAAA----CGGUCACACUAAUAUGAA-- (((.(((((..((((..((..(.(((..........))).)..))..))))----))))).))).........-- ( -17.70, z-score = -0.78, R) >droSec1.super_1 11804740 65 + 14215200 GUGCGACCGCAUUUGGAUGGCGGAAAAAUGGUGACAUU----UCAUGCAUG----UGGUCACACUAAUAUGAA-- (((.((((((((.......(((...(((((....))))----)..))))))----))))).))).........-- ( -22.81, z-score = -3.04, R) >droSim1.chr2R 13034390 65 + 19596830 GUGCGACCGCAUUUGGAUGGAGGAAAAAUGGUGACAUU----UCAAGCAUG----UGGUCACACUAAUAUGAA-- (((.(((((((((((((..........(((....))))----))))..)))----))))).))).........-- ( -20.00, z-score = -2.69, R) >consensus GUGCGACCGCAUUUGGAUGGAGGGAAAAUGGUGAAAUU____UCACGCAAA____UGGUCACACUAAUAUGAA__ (((.((((((((((...........)))))((((........)))).........))))).)))........... ( -9.72 = -9.80 + 0.08)
| Location | 14,327,989 – 14,328,056 |
|---|---|
| Length | 67 |
| Sequences | 5 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Shannon entropy | 0.38956 |
| G+C content | 0.42561 |
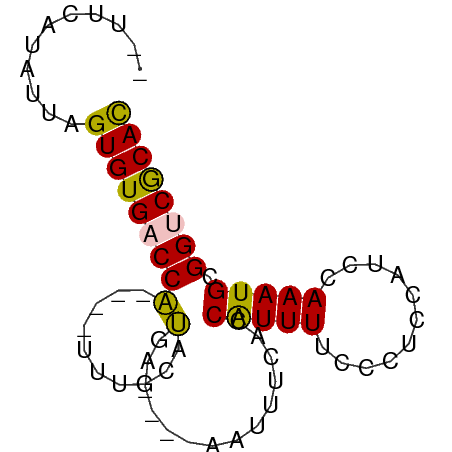
| Mean single sequence MFE | -14.22 |
| Consensus MFE | -7.73 |
| Energy contribution | -7.81 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14327989 67 - 21146708 UGUUCAUAUUAGUGUGACCA----UUUGCAUAA----AAUUUCACCGUUUUCCCUCAAUCCAAAUGCGGCCGCAU ...........(((((.(((----((((.((((----(((......)))))......)).)))))).)..))))) ( -6.60, z-score = 0.81, R) >droEre2.scaffold_4845 8535845 73 - 22589142 --UUUUUAUUAGUGUGACCGUUUCUUUGUGCGCAAUAAAUUUUGCCAUUUUCCCACUAUCAAAAUGCGGUCACAC --.........((((((((((...((((((.((((......))))))............))))..)))))))))) ( -20.60, z-score = -4.56, R) >droYak2.chr2R 6280766 69 + 21139217 --UUCAUAUUAGUGUGACCG----UUUGCGCGAAACGAAUUUAACCAUUUUCCCCCCAUCCAAAUGCGGCCGCAC --.........(((((.(((----((((........(((..........)))........)))..)))).))))) ( -10.29, z-score = -0.13, R) >droSec1.super_1 11804740 65 - 14215200 --UUCAUAUUAGUGUGACCA----CAUGCAUGA----AAUGUCACCAUUUUUCCGCCAUCCAAAUGCGGUCGCAC --.........((((((((.----(((((..((----((((....))))))...)).......))).)))))))) ( -17.61, z-score = -2.69, R) >droSim1.chr2R 13034390 65 - 19596830 --UUCAUAUUAGUGUGACCA----CAUGCUUGA----AAUGUCACCAUUUUUCCUCCAUCCAAAUGCGGUCGCAC --.........((((((((.----(((....((----((((....))))))............))).)))))))) ( -15.99, z-score = -2.82, R) >consensus __UUCAUAUUAGUGUGACCA____UUUGCAUGA____AAUUUCACCAUUUUCCCUCCAUCCAAAUGCGGUCGCAC ...........((((((((................................................)))))))) ( -7.73 = -7.81 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:05 2011