
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,323,128 – 14,323,228 |
| Length | 100 |
| Max. P | 0.548470 |

| Location | 14,323,128 – 14,323,228 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 65.21 |
| Shannon entropy | 0.70019 |
| G+C content | 0.47656 |
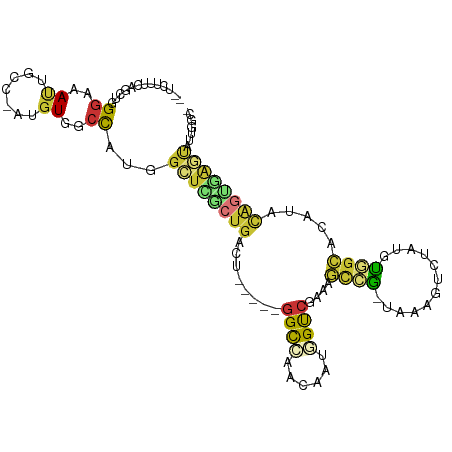
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.59 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.95 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
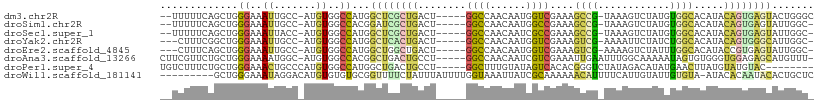
>dm3.chr2R 14323128 100 + 21146708 --UUUUUCAGCUGGGAAAUUGCC-AUGUGGCCAUGGCUCGCUGACU-----GGCCAACAAUGGUCGAAAGCCG-UAAAGUCUAUGUGGCACAUACAGUGAGUACUGGGC --.......(((.((.....(((-(((....))))))((((((..(-----(((((.((((((.(....))))-)........)))))).))..))))))...)).))) ( -32.80, z-score = -0.88, R) >droSim1.chr2R 13029555 99 + 19596830 --UUUUUCAGCUGGGAAAUUGCC-AUGUGGCCACGGAUCGCUGACU-----GGCCAACAAUGGCCGAAAGCCG-UAAAGUCUAUGUGGCACAUACAGUGAGUAUUGGC- --...((((.(((......((((-((((((..((..((.(((...(-----(((((....))))))..))).)-)...))))))))))))....))))))).......- ( -30.90, z-score = -0.66, R) >droSec1.super_1 11799914 99 + 14215200 --UUUUUCAGCUGGGAAAUUACC-AUGUGGCCAUGGCUCGCUGACU-----GGCCAACAAUCGCCGAAAGCCG-UAAAGUCUAUGUGGCACAUACAGUGAGUAUUGGC- --.......(((((.......))-).)).((((..((((((((..(-----(((........))))...((((-((.......)))))).....))))))))..))))- ( -29.60, z-score = -0.61, R) >droYak2.chr2R 6275643 98 - 21139217 ---CUUUCGGCUGGGAAAUUGCC-AUGUGGCCAUGGCUCACUGACU-----GGCCAACAAUGGUCGAAAGUCG-AAAAUUCUAUCUGGCACAUACAGUGGGCAUUGGC- ---.....(((.........)))-.....((((..((((((((..(-----(((((..(((..(((.....))-)..))).....)))).))..))))))))..))))- ( -32.50, z-score = -1.78, R) >droEre2.scaffold_4845 8530997 98 + 22589142 ---CUUUCAGCUGGGAAAUUGCC-AUGUGGCCAUGGCUGGCUGACU-----GGCCAACAAUGGUCGAAAGUCG-AAAAGUCUAUUUGGCACAUACCGUGAGUAUUGGC- ---......((..(....(..(.-.((((((((.((((...(((((-----(((((....)))))...)))))-...))))....)))).))))..)..)...)..))- ( -29.60, z-score = -0.63, R) >droAna3.scaffold_13266 15676781 102 - 19884421 CUUCGUUCUGCUGGGAAAAUGGC-AUGUGGCCACGGCUGACUGCCU-----GGCCAACAAUCGUCGAAAUUGAAUUUGGCAAAAAUAGUGUGGGUGGAGAGCAUGUUU- (((((..((((((..........-.((((((((.(((.....))))-----)))).)))...((((((......)))))).....))))).)..))))).........- ( -31.30, z-score = -1.01, R) >droPer1.super_4 6977725 96 + 7162766 UGUCUUUCUGCUGGGAAACUGCCCAUGUGGCCAUGGCUGACUGCCU-----GGCUUUGUAUAGUCACACGGGUCUAUAGACAUAUGAACUUAUGUAUGUAC-------- .........((((((......)))).))(((((.(((.....))))-----))))..(((((...(((..(((.((((....)))).)))..))).)))))-------- ( -26.90, z-score = -0.66, R) >droWil1.scaffold_181141 1919584 99 - 5303230 ---------GCUGGGAAAUAGGACAUGUGUGUGCGGUUUUCUAUUUAUUUUGGUAAAUUAUCGCAAAAAACAUUUUCAUUGUAUUGUGUA-AUACACAAUACACUGCUC ---------((.((......(((.((((...((((((.....((((((....)))))).))))))....)))).)))..(((((((((..-...))))))))))))).. ( -25.90, z-score = -2.99, R) >consensus __UCUUUCAGCUGGGAAAUUGCC_AUGUGGCCAUGGCUCGCUGACU_____GGCCAACAAUGGUCGAAAGCCG_UAAAGUCUAUGUGGCACAUACAGUGAGUAUUGGC_ .............((..((.......))..))...((((((((........((((......))))....((((............)))).....))))))))....... (-10.68 = -10.59 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:04 2011