
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,214,312 – 3,214,414 |
| Length | 102 |
| Max. P | 0.984254 |

| Location | 3,214,312 – 3,214,414 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.66 |
| Shannon entropy | 0.67106 |
| G+C content | 0.45923 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -14.37 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3214312 102 + 23011544 GGUUCUGGGUG-----CCGGGACUU-UCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG- ((((((((...-----)))))))).-..((((.((((((((....))))))))....((((..((((((....)))))))))).))))((((((....))))))....- ( -34.10, z-score = -2.05, R) >droSim1.chr2L 3153453 102 + 22036055 GGUUCUCGGGG-----CCGGGACUU-CCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG- .....((((((-----.........-)))))).((((((((....))))))))....((((..((((((....))))))))))(((((((....))))))).......- ( -34.60, z-score = -1.78, R) >droSec1.super_5 1363913 102 + 5866729 GGUUCUGGGGG-----CCGGGACUU-CCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG- ....(..((((-----(((((....-.)))...((((((((....))))))))...))).....(....)......)))..).(((((((....))))))).......- ( -34.30, z-score = -1.54, R) >droYak2.chr2L 3203405 107 + 22324452 GGUUUUGGGGGAGGGACCGGGACUU-CCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG- ...((((((((((.........)))-)))))))((((((((....))))))))....((((..((((((....))))))))))(((((((....))))))).......- ( -40.30, z-score = -3.00, R) >droEre2.scaffold_4929 3251492 88 + 26641161 -------------------GGUUUU-CCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG- -------------------(((.((-.(((((.((((((((....))))))))....((((..((((((....)))))))))).))))).))..)))...........- ( -26.50, z-score = -1.96, R) >droAna3.scaffold_12904 24028 109 - 61084 GAUGGCGGCGGCGAGGAAAGGACAUGCCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUGC .....(((((.((((((((......(((.....((((((((....))))))))...))).....(....)...)))))))).))))).((((((....))))))..... ( -38.00, z-score = -2.29, R) >dp4.chr4_group4 1289346 99 + 6586962 ------CGCAG----AGAGCUACAGCAGCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGAAAACACAUUUUCUUGAUGUCGAGCAAUCGCCUGAUUGUAUUGC ------.((((----.((((....))(((((..((((((((....))))))))..)))))..(((((((....)))))))....))..((((((....)))))).)))) ( -26.30, z-score = -1.45, R) >droPer1.super_10 3034674 99 - 3432795 ------CGCAG----AGAGCAACAGCAACCGAAGGCACUGAAAUAUUAAUGCUAAUAGCUAGAAGGAUACACAUUUCCAUGGUGUCGGGCAAUCGCCAGAUUGUAUUGC ------.((..----...))....((((((((.((((.(((....))).))))....((((...(((........))).)))).))))((((((....)))))).)))) ( -24.00, z-score = -0.82, R) >droWil1.scaffold_180708 545430 100 - 12563649 ------ACCAGCCAAGGCUAGGCGAGCCUUCAAGGUUUUGAAAUAUUAAUGCUAAUGGCUAAAAGGGAAU-CACUUUUUUGCUUUAGCCUUAACUUGUAUUGCCUUU-- ------.......(((((...(((((..(((((....)))))...((((.(((((.(((.((((((....-..)))))).)))))))).)))))))))...))))).-- ( -26.80, z-score = -0.85, R) >droGri2.scaffold_15252 7869649 80 - 17193109 -----AGUCAGAGCCGACCAGACGC-AAUCCAAUCCAUUGAAAUAUUAAUGCUAAUGGCUAAAGGGAAACAUUUGCUGAUUGUGUU----------------------- -----.(((......)))..(((((-((((.....((((((....)))))).....(((.(((.(....).)))))))))))))))----------------------- ( -18.10, z-score = -1.16, R) >consensus ______GGCAG_____CCGGGACUU_CCCCGAAGGCAUUGAAAUAUUAAUGCUAAUGGCUAGAAGGAAACACAUUUCCUUGGCGUCGGGCAAUCGCCUGAUUGUAUUG_ .................................((((((((....))))))))....((((..((((((....))))))))))(((((((....)))))))........ (-14.37 = -15.94 + 1.57)

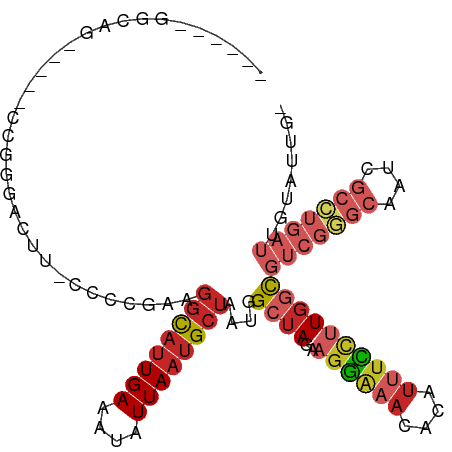
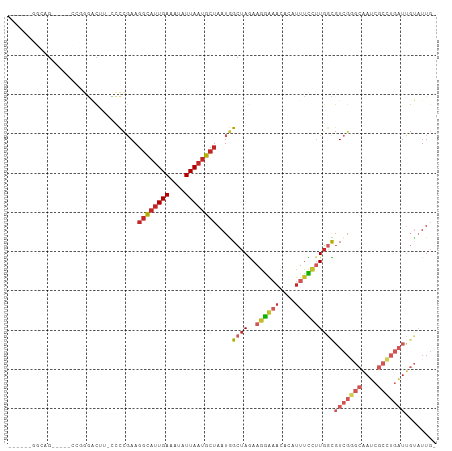
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:18 2011