
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,313,917 – 14,314,011 |
| Length | 94 |
| Max. P | 0.583761 |

| Location | 14,313,917 – 14,314,011 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.65552 |
| G+C content | 0.40346 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -9.63 |
| Energy contribution | -11.56 |
| Covariance contribution | 1.93 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14313917 94 + 21146708 GUUUG--CUUUGACGAGGUGGCAACGCCAUAGAGGG-UUCUUGCAUACUUCUCAAUUUUUUCUUCAGGCCACAAGAAAAUCUUUAGAUAUGAGGAAU .....--..((((.((((((((((.(((......))-)..)))).))))))))))....(((((((......(((.....)))......))))))). ( -22.70, z-score = -0.89, R) >droAna3.scaffold_13266 15665081 89 - 19884421 GUUCAAAGUAAAACAGUGUGGCAACGCCGC-------UCCAUUCAU-UUUCCUAAAUUUUUCUUUGAUUCCUAAGCGAAUUAUGGAAAAAUGGAAAU ...............(.(((((...)))))-------.).......-(((((...((((((((.((((((......)))))).))))))))))))). ( -20.00, z-score = -2.23, R) >droEre2.scaffold_4845 8521836 94 + 22589142 GCUUG--GUUUGGCGAGGUGGCAACGCCACCAAGGG-UUCUUGCAUAUUCCUCAGUUUUUUCUUCAGGCCAAUAGAAAAUCUUUAGAUAUGAGGAAU ..(((--((((((.((((((((...)))))).((((-...........))))........)).))))))))).......((((((....)))))).. ( -26.20, z-score = -1.09, R) >droYak2.chr2R 6266372 94 - 21139217 GCUUG--CUUUGGCGAGGUGGCAACACCACAAAUGG-UUCUUAUAUAUUUCUCAGUUUUUUCUUCAGCCCACAAGAAAAUCUUUAGAUAUGACGAAU ..(((--(....))))((((....)))).......(-((((((((....(((.((..(((((((........)))))))..)).)))))))).)))) ( -19.10, z-score = -0.88, R) >droSec1.super_1 11790752 95 + 14215200 GGUUG--CUUUGACGAGGUGGCAACGCCAUAGGGGGGUUCUUGCAUAUUUCUCAGUUUUUUCUUCAGGCCACAAGAAAAUCUUUAGAUAUGAGGAAU .((.(--(((.((.((((((....))))...(((..((........))..))).......)).)))))).)).......((((((....)))))).. ( -24.30, z-score = -0.81, R) >droSim1.chr2R 13050678 94 + 19596830 GGUUG--CUUUCACGAGGUGGCAACGCCAUAAAGGG-UUCUUGCAUAUUUCUCAGUUUUUUCUUCAGGCCACAAGAAAAUCUUUAGAUAUGAGGAAU .((((--((.((....)).)))))).((.....))(-(((((.(((((((...((..(((((((........))))))).))..))))))))))))) ( -21.20, z-score = -0.61, R) >droVir3.scaffold_12875 650943 84 - 20611582 GCUAA--GCAGAACAGC-UGGCAACGCUCGG-------CAGUACAAUUUUUAUAAGUGAC-CGACAGAAAAGCCGUAAAUGA--AAAAAUGACAAAC ((((.--((......))-))))......(((-------(.((...((((....)))).))-(....)....)))).......--............. ( -10.50, z-score = 1.16, R) >consensus GCUUG__CUUUGACGAGGUGGCAACGCCACAAAGGG_UUCUUGCAUAUUUCUCAGUUUUUUCUUCAGGCCACAAGAAAAUCUUUAGAUAUGAGGAAU ................((((....)))).........(((((.(((((((.......(((((((........))))))).....)))))))))))). ( -9.63 = -11.56 + 1.93)
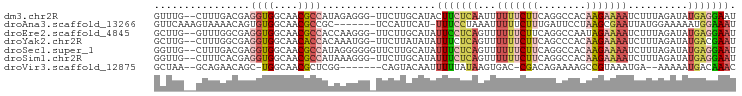
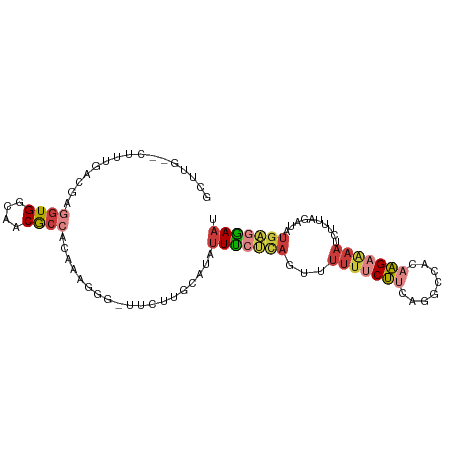

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:02 2011