
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,219,467 – 14,219,538 |
| Length | 71 |
| Max. P | 0.541002 |

| Location | 14,219,467 – 14,219,538 |
|---|---|
| Length | 71 |
| Sequences | 10 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.59149 |
| G+C content | 0.45295 |
| Mean single sequence MFE | -8.88 |
| Consensus MFE | -4.63 |
| Energy contribution | -4.49 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14219467 71 - 21146708 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAA----AGCCAACCGACCGCACUAUUCCG -..................((((..-.((((.....))))..........----.(.....)))))........... ( -7.60, z-score = -0.47, R) >droYak2.chr2R 6177555 71 + 21139217 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAA----AGCCAGCCGACCGCACUAUUCCG -..................((((..-.((((.....))))..........----.(....).))))........... ( -8.40, z-score = -0.38, R) >droSec1.super_1 11708841 71 - 14215200 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAA----AGCCAACCGACCGCACUAUUCCG -..................((((..-.((((.....))))..........----.(.....)))))........... ( -7.60, z-score = -0.47, R) >droSim1.chr2R 12956851 71 - 19596830 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAA----AGCCAACCGACCGCACUAUUCCG -..................((((..-.((((.....))))..........----.(.....)))))........... ( -7.60, z-score = -0.47, R) >droVir3.scaffold_12875 560978 68 + 20611582 GCGCAC-CUCCGCAUAUUUUGUUGUAUAUGGUAUCUAUAUUUAUGCAUACAUUUGAACAGACUGUCUCG-------- (((...-...)))(((.((((((.((....((((.(((....))).))))...)))))))).)))....-------- ( -7.30, z-score = 1.22, R) >droEre2.scaffold_4845 8440568 57 - 22589142 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACCG----AGCCAACCG-------------- -....(((....(((.....)))..-.((((.....)))).........)----)).......-------------- ( -7.80, z-score = -1.06, R) >droWil1.scaffold_181141 1730836 56 + 5303230 --GCGCGCCCCAGACAUUUGGUCAC-CAUGCAAUUUGCAUAAAUUAAC------AGACAAGCGAC------------ --.(((......(((.....)))..-.((((.....))))........------......)))..------------ ( -10.00, z-score = -0.88, R) >droAna3.scaffold_13266 15577680 72 + 19884421 -UUCACUCCCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAAAUAGAAUAGCCAGCCAGCUGUUCUAUU--- -...........(((.....)))..-.((((.....))))......(((((((((((......)))))))))))--- ( -17.90, z-score = -3.63, R) >dp4.chr3 11496023 59 - 19779522 -UUCACUCGCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAG----AACCCCCUACG------------ -(((....(((((....)))))...-.((((.....)))).........)----)).........------------ ( -7.30, z-score = -0.78, R) >droPer1.super_4 6899626 59 - 7162766 -UUCACUCGCCAGACAUUUGGUCAC-CGUGCAAUUUGCAUAAAUUAACAG----AACCCCCUACG------------ -(((....(((((....)))))...-.((((.....)))).........)----)).........------------ ( -7.30, z-score = -0.78, R) >consensus _UUCACUCCCCAGACAUUUGGUCAC_CGUGCAAUUUGCAUAAAUUAACAA____AGCCAACCGACCGC_________ ............(((.....)))....((((.....))))..................................... ( -4.63 = -4.49 + -0.14)

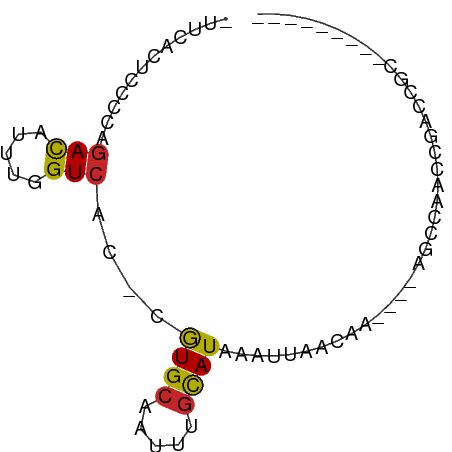

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:52 2011