
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,179,907 – 14,180,054 |
| Length | 147 |
| Max. P | 0.938083 |

| Location | 14,179,907 – 14,180,054 |
|---|---|
| Length | 147 |
| Sequences | 7 |
| Columns | 151 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.50365 |
| G+C content | 0.51026 |
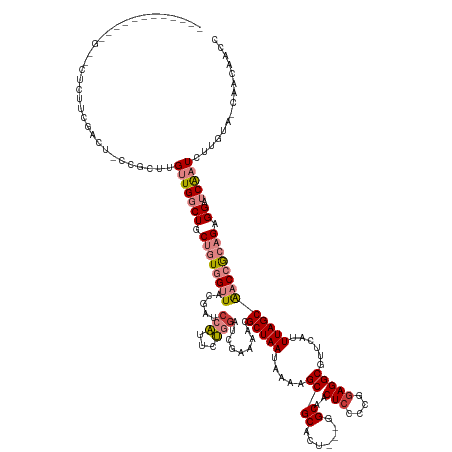
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -19.15 |
| Energy contribution | -21.07 |
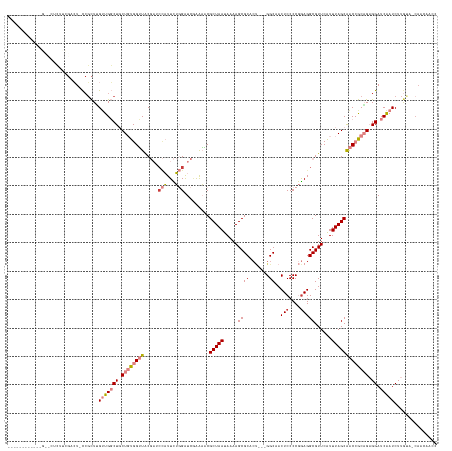
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
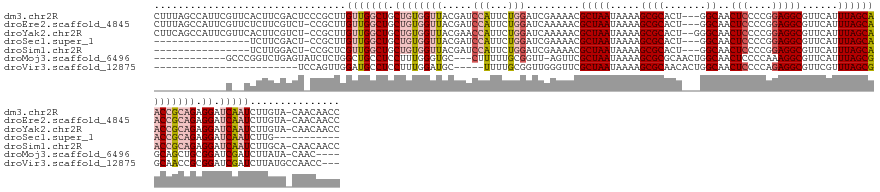
>dm3.chr2R 14179907 147 - 21146708 CUUUAGCCAUUCGUUCACUUCGACUCCCGCUUGUUGGCUGCUGUGGUUACGAUCCAUUCUGGAUCGAAAACGCUAAUAAAAGCGCACU---GGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUGUA-CAACAACC ....(((...(((.......))).....)))(((((....((((((((.((((((.....)))))).....(((((....(((((.((---((.......))))..)))))...)))))))))))))((((....))))...-)))))... ( -43.90, z-score = -2.20, R) >droEre2.scaffold_4845 8400770 146 - 22589142 CUUUAGCCAUUCGUUCUCUUCGUCU-CCGCUUGUUGGCUGCUGUGGUUACGAUCCAUUCUGGAUCAAAAACGCUAAUAAAAGCGCACU---GGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUGUA-CAACAACC ....(((....((.......))...-..)))(((((....((((((((..(((((.....)))))......(((((....(((((.((---((.......))))..)))))...)))))))))))))((((....))))...-)))))... ( -41.30, z-score = -1.90, R) >droYak2.chr2R 6137726 147 + 21139217 CUUCAGCCAUUCGUUCACUUCGUCU-CCGCUUGUUGGCUGCUGUGGUUACGAACCAUUCUGGAUCAAAAACGCUAAUAAAAGCGCACU--GGGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUGUA-CAACAACC ...((((((................-........))))))((((((((..((.((.....)).))......(((((....(((((.((--(((......)))))..)))))...)))))))))))))((((....))))...-........ ( -41.56, z-score = -1.46, R) >droSec1.super_1 11669756 120 - 14215200 ----------------UCUUCGACU-CCGCUUGUUGGCUGCUGUGGUUACGAUCCAUUCUGGAUCGAAAACGCUAAUAAAAGCGCACU---GGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUG----------- ----------------.....((.(-(((((....)))..((((((((.((((((.....)))))).....(((((....(((((.((---((.......))))..)))))...))))))))))))).))))).......----------- ( -43.70, z-score = -3.55, R) >droSim1.chr2R 12916336 130 - 19596830 ----------------UCUUGGACU-CCGCUCGUUGGCUGCUGUGGUUACGAUCCAUUCUGGAUCGAAAACGCUAAUAAAAGCGCACU---GGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUGCA-CAACAACC ----------------.........-..((..(((((((.((((((((.((((((.....)))))).....(((((....(((((.((---((.......))))..)))))...))))))))))))).)).)))))...)).-........ ( -44.20, z-score = -2.93, R) >droMoj3.scaffold_6496 6080608 130 - 26866924 ------------GCCCGGUCUGAGUAUCUCUGGCUGCCUCCUUUGGGUGC---CUUUUUGCGGUU-AGUUCGCUAAUAAAAGCGCGCAACUGGCAACUCCCCAAAGGCGUUCAUUUAGCGGCAGCUGCGGAUCGAUCUUAUA-CAAC---- ------------...(((((((((.....))(((((((.(((((((((((---(...(((((...-....((((......)))))))))..))))....)))))))).(((.....)))))))))).)))))))........-....---- ( -47.11, z-score = -2.20, R) >droVir3.scaffold_12875 509943 119 + 20611582 ------------------------UCCAGUUGGAUGCCUCCUUUGGAUGC-----UUUUGCGGUUGGGUUCGCUAAUAAAAGCGCAACACUGGCAACUCCCCAGAGGCGUUCGUUUAGCGGCAACCGCGGAUCGAUCUUAUGCCAACC--- ------------------------....(((((..((.(((...))).))-----.((((((((((...(((((((....(((((....((((.......))))..)))))...)))))))))))))))))...........))))).--- ( -41.30, z-score = -1.43, R) >consensus ____________G__CUCUUCGACU_CCGCUUGUUGGCUGCUGUGGUUACGAUCCAUUCUGGAUCGAAAACGCUAAUAAAAGCGCACU___GGCAACUCCCCGGAGGCGUUCAUUUAGCAACCGCAGAGGAUCAAUCUUGUA_CAACAACC ................................(((((((.((((((((..((.(((...))).))......(((((....(((((......(....)((....)).)))))...))))))))))))).)).)))))............... (-19.15 = -21.07 + 1.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:49 2011