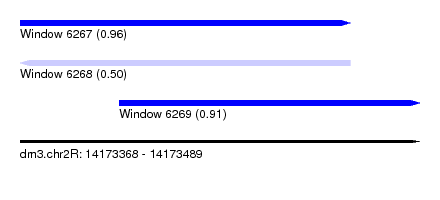
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,173,368 – 14,173,489 |
| Length | 121 |
| Max. P | 0.957631 |

| Location | 14,173,368 – 14,173,468 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.50167 |
| G+C content | 0.68085 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -25.73 |
| Energy contribution | -24.72 |
| Covariance contribution | -1.01 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14173368 100 + 21146708 CCGGUCGCUGCCAGAAGUGAGGCAGCGGCAGGCACCACGGUCGAUGGUGAACCGGUGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG ((.(((((((((........))))))))).....(((((((((....)).))).))))..))((.(((.((((..(((((....)))))))))))).)). ( -43.20, z-score = -1.42, R) >droGri2.scaffold_15245 11428529 85 + 18325388 GUUGUAGCAGCCAGCAGCGAUGCUGCAGCAGAGACAACAGCAGCUGG---------------UGAGCCAGCAGAGGGCGUAAAUACGCCUGGUGGCACAG (((((..(.((..(((((...))))).)).)..))))).((.(((((---------------....)))))...((((((....))))))....)).... ( -35.50, z-score = -2.16, R) >droVir3.scaffold_12875 502954 85 - 20611582 GUCGUUGCAGCCAGCAGCGAGGCAGCCGCCGAGACAACAGCUGCCGG---------------GGAGCCAGCCGAGGGCGUGAAUACGCCCGGCGGCACAG .(((((((.....)))))))((((((.(.........).))))))((---------------....)).((((.((((((....))))))..)))).... ( -42.70, z-score = -2.85, R) >droPer1.super_4 6853555 100 + 7162766 GUUGUGGCUGCCAGCAGAGAGGCGGCUGCUGGGACGACGGCCGAUGGAGAUCCCGCCGAGGGUGAGCCGGCCGACGGUGUAAACACCCCCGGCGGCACAG ..((((((((((........))))))(((((((....(((((...((....(((.....)))....)))))))..((((....))))))))))).)))). ( -48.40, z-score = -1.84, R) >dp4.chr3 11449302 100 + 19779522 GUUGUGGCUGCCAGCAGAGAGGCGGCUGCUGGGACGACGGCCGAUGGAGAUCCCGCCGAGGGUGAGCCGGCCGACGGUGUAAACACCCCCGGCGGCACAG ..((((((((((........))))))(((((((....(((((...((....(((.....)))....)))))))..((((....))))))))))).)))). ( -48.40, z-score = -1.84, R) >droSim1.chr2R 12909916 100 + 19596830 CCGGUCGCCGCCAGAAGUGAGGCAGCGGCAGGCACCACGGUCGAUGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG .((((.((((((...((((.(((..(((....((((((....).)))))..))).(....)....)))))))...(((((....))))).)))))))))) ( -42.90, z-score = -1.05, R) >droSec1.super_1 11663273 100 + 14215200 CCGGUCGCCGCCAGAAGUGAGGCAGCGGCAGGCACCACGGUCGAUGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG .((((.((((((...((((.(((..(((....((((((....).)))))..))).(....)....)))))))...(((((....))))).)))))))))) ( -42.90, z-score = -1.05, R) >droYak2.chr2R 6131306 100 - 21139217 CCGGUCGCCGCCAGAAGUGAGGCAGCCGCAGGCACCACGUUCGAAGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG .((((.((((((...((((.(((.(((...))).((...((((..((....))..)))).))...)))))))...(((((....))))).)))))))))) ( -44.30, z-score = -1.92, R) >droEre2.scaffold_4845 8394419 100 + 22589142 CCGGUCGCCGCCAGAAGUGAGGCAGCCGCAGGCACCACGGUCGACGGUGAGCCGGCGGAUGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG .((((.((((((...((((.(((..((((.(((..(((.(....).))).))).)))).......)))))))...(((((....))))).)))))))))) ( -46.10, z-score = -1.45, R) >consensus CCGGUCGCCGCCAGAAGUGAGGCAGCCGCAGGCACCACGGUCGAUGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUG .((((.((((((..........((((.((.(......).))....((....((.......))....)).))))..(((((....))))).)))))))))) (-25.73 = -24.72 + -1.01)
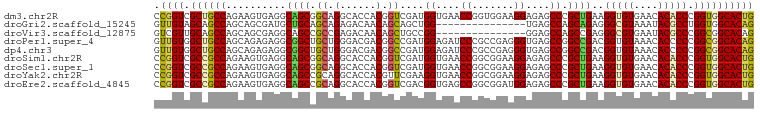
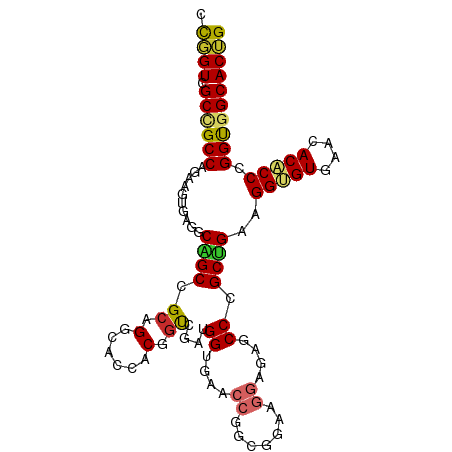
| Location | 14,173,368 – 14,173,468 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.50167 |
| G+C content | 0.68085 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 14173368 100 - 21146708 CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCUUCCACCGGUUCACCAUCGACCGUGGUGCCUGCCGCUGCCUCACUUCUGGCAGCGACCGG ........((((((((....)))))....((((((.......((((.((((.......)))))))).))))))(((((((........)))))))..))) ( -41.80, z-score = -2.37, R) >droGri2.scaffold_15245 11428529 85 - 18325388 CUGUGCCACCAGGCGUAUUUACGCCCUCUGCUGGCUCA---------------CCAGCUGCUGUUGUCUCUGCUGCAGCAUCGCUGCUGGCUGCUACAAC .((.((((.(((((((....)))))...)).)))).))---------------(((((.((((((((.......))))))..)).))))).......... ( -30.20, z-score = -0.94, R) >droVir3.scaffold_12875 502954 85 + 20611582 CUGUGCCGCCGGGCGUAUUCACGCCCUCGGCUGGCUCC---------------CCGGCAGCUGUUGUCUCGGCGGCUGCCUCGCUGCUGGCUGCAACGAC ....((((((((((((....))))))..))).)))...---------------..(((((((((((...)))))))))))(((.(((.....))).))). ( -43.40, z-score = -2.48, R) >droPer1.super_4 6853555 100 - 7162766 CUGUGCCGCCGGGGGUGUUUACACCGUCGGCCGGCUCACCCUCGGCGGGAUCUCCAUCGGCCGUCGUCCCAGCAGCCGCCUCUCUGCUGGCAGCCACAAC ..((.((((((((((((......(((.....)))..)))))))))))).)).......(((.(.....(((((((........)))))))).)))..... ( -43.30, z-score = -1.04, R) >dp4.chr3 11449302 100 - 19779522 CUGUGCCGCCGGGGGUGUUUACACCGUCGGCCGGCUCACCCUCGGCGGGAUCUCCAUCGGCCGUCGUCCCAGCAGCCGCCUCUCUGCUGGCAGCCACAAC ..((.((((((((((((......(((.....)))..)))))))))))).)).......(((.(.....(((((((........)))))))).)))..... ( -43.30, z-score = -1.04, R) >droSim1.chr2R 12909916 100 - 19596830 CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCUUCCGCCGGUUCACCAUCGACCGUGGUGCCUGCCGCUGCCUCACUUCUGGCGGCGACCGG ........((((((((....)))))....((((((.......((((.((((.......)))))))).))))))(((((((........)))))))..))) ( -40.70, z-score = -1.26, R) >droSec1.super_1 11663273 100 - 14215200 CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCUUCCGCCGGUUCACCAUCGACCGUGGUGCCUGCCGCUGCCUCACUUCUGGCGGCGACCGG ........((((((((....)))))....((((((.......((((.((((.......)))))))).))))))(((((((........)))))))..))) ( -40.70, z-score = -1.26, R) >droYak2.chr2R 6131306 100 + 21139217 CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCUUCCGCCGGUUCACCUUCGAACGUGGUGCCUGCGGCUGCCUCACUUCUGGCGGCGACCGG ........((((((((....))))).((.((((((.......((((..((((......)))))))).)))))).((((((........)))))))).))) ( -39.10, z-score = -0.65, R) >droEre2.scaffold_4845 8394419 100 - 22589142 CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCAUCCGCCGGCUCACCGUCGACCGUGGUGCCUGCGGCUGCCUCACUUCUGGCGGCGACCGG ........((((((((....))))).((.((((((...((((.((.(((....))).))...)))).)))))).((((((........)))))))).))) ( -42.30, z-score = -0.66, R) >consensus CAGUGCCACCGGGUGUGUUCACACCUUCAGCGGGCUCUCCUUCCGCCGGUUCACCAUCGACCGUGGUGCCUGCCGCUGCCUCACUUCUGGCGGCGACCGG .((.(((..(.(((((....)))))....)..))).)).....((((((.((......)).).)))))......((((((........))))))...... (-20.00 = -21.08 + 1.08)
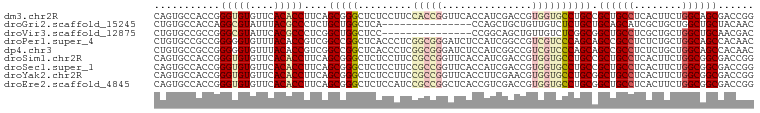
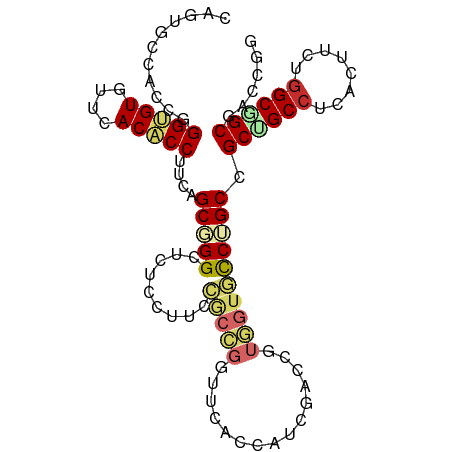
| Location | 14,173,398 – 14,173,489 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.36 |
| Shannon entropy | 0.71424 |
| G+C content | 0.65475 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14173398 91 + 21146708 GGCACCACGGUCGAUGGUGAACCGGUGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUGCUGACAUGGAGGAUG-AACCAC------------------ ((((((((....).)))))..((.(((..((.((.(((.((((..(((((....)))))))))))).)).))..)))...))...-..))..------------------ ( -31.80, z-score = -0.73, R) >droGri2.scaffold_15245 11428559 72 + 18325388 ---------------GAGACAACAGCAGCUGGUGAGCCAGCAGAGGGCGUAAAUACGCCUGGUGGCACAGCGGAUGCAGAGGAAUUG----------------------- ---------------.........(((.(((.((.((((.((...(((((....))))))).)))).)).))).)))..........----------------------- ( -26.90, z-score = -3.02, R) >droMoj3.scaffold_6496 6073348 72 + 26866924 ---------------GAGACCACAGCAGCCGGCGAGCCGGCAGAGGGCGUAAAUACGCCGGGCGGCACAGCCGUGGCUGUGGCAUUG----------------------- ---------------..(.(((((((.(((((....)))))....(((((....)))))..(((((...))))).))))))))....----------------------- ( -38.90, z-score = -3.82, R) >droVir3.scaffold_12875 502984 72 - 20611582 ---------------GAGACAACAGCUGCCGGGGAGCCAGCCGAGGGCGUGAAUACGCCCGGCGGCACAGCCGUUGCGCUCGAGCUG----------------------- ---------------(((.((((.((((..((....)).((((.((((((....))))))..)))).)))).))))..)))......----------------------- ( -35.70, z-score = -2.28, R) >droAna3.scaffold_13266 15534900 91 - 19884421 GGCACAACAGCCGAUGGCGACCCACCGGAUGGGGAUCCAGCCGACGGCGUGAACACCCCCGGGGGCACAGCGGACAUGGAGGAGC-UACUGU------------------ (((.(..((.(((.((..(.(((.((((..((((.((..(((...)))..)).).))))))))))).)).)))...))...).))-).....------------------ ( -31.20, z-score = 0.62, R) >droPer1.super_4 6853585 104 + 7162766 GGGACGACGGCCGAUGGAGAUCCCGCCGAGGGUGAGCCGGCCGACGGUGUAAACACCCCCGGCGGCACAGCUGACAUGGAGGAGGCAGCUGCAGCUGCAGCAGC------ (((((..((.....))..).))))((((.(((((.((((.....)))).....))))).)))).((...(((..(.....)..))).(((((....))))).))------ ( -44.60, z-score = -1.01, R) >dp4.chr3 11449332 110 + 19779522 GGGACGACGGCCGAUGGAGAUCCCGCCGAGGGUGAGCCGGCCGACGGUGUAAACACCCCCGGCGGCACAGCUGACAUGGAGGAGGCAGCUGCAGCUGCAGCUGCAGCAGC (((((..((.....))..).))))((((.(((((.((((.....)))).....))))).)))).((...(((..(.....)..))).((((((((....)))))))).)) ( -52.10, z-score = -2.07, R) >droWil1.scaffold_181141 1675111 76 - 5303230 ---------------GACACCACAGCUGCCGGUGAACCAGCCGAUGGUGUAAACACUCCCGGAGGCACCGCCGACAUGGAGGCAGAGGCAG------------------- ---------------..........(((((((((..((..(((..((((....))))..))).))))))(((........)))...)))))------------------- ( -28.60, z-score = -1.78, R) >droSim1.chr2R 12909946 91 + 19596830 GGCACCACGGUCGAUGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUGCUGACAUGGAGGAUG-AACCAC------------------ ....(((..((((..((((.((((((((.........))))....(((((....)))))))))..))))..)))).))).((...-..))..------------------ ( -32.10, z-score = -0.81, R) >droSec1.super_1 11663303 91 + 14215200 GGCACCACGGUCGAUGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUGCUGACAUGGAGGAUG-AACCAC------------------ ....(((..((((..((((.((((((((.........))))....(((((....)))))))))..))))..)))).))).((...-..))..------------------ ( -32.10, z-score = -0.81, R) >droYak2.chr2R 6131336 91 - 21139217 GGCACCACGUUCGAAGGUGAACCGGCGGAAGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUGCUGACAUGGAGGAUG-AGCCAC------------------ (((....((((((..((....))..)(..((.((.(((.((((..(((((....)))))))))))).)).))..).....)))))-.)))..------------------ ( -32.20, z-score = -0.87, R) >droEre2.scaffold_4845 8394449 91 + 22589142 GGCACCACGGUCGACGGUGAGCCGGCGGAUGGAGAGCCCGCUGAAGGUGUGAACACACCCGGUGGCACUGCAGACAUGGAGGAUG-AACCAC------------------ ....(((..(((..(((((.((((((((.........))))....(((((....)))))))))..)))))..))).))).((...-..))..------------------ ( -33.90, z-score = -0.84, R) >consensus GGCACCACGGUCGAUGGUGAACCAGCGGAAGGAGAGCCCGCCGAAGGUGUGAACACACCCGGUGGCACAGCUGACAUGGAGGAUG_AACCAC__________________ .............................((.((.(((.((((..(((((....)))))))))))).)).))...................................... (-13.63 = -13.98 + 0.36)
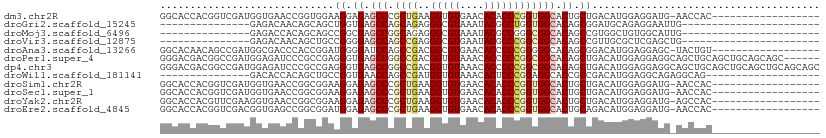

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:47 2011