
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,167,614 – 14,167,696 |
| Length | 82 |
| Max. P | 0.545977 |

| Location | 14,167,614 – 14,167,696 |
|---|---|
| Length | 82 |
| Sequences | 14 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.51040 |
| G+C content | 0.57431 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -17.72 |
| Energy contribution | -18.19 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.545977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
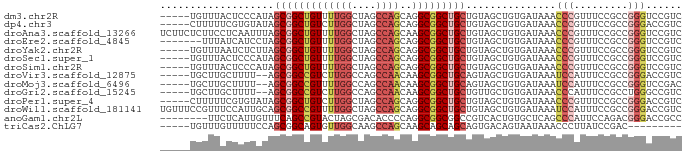
>dm3.chr2R 14167614 82 - 21146708 -----UGUUUACUCCCAUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC -----...((((..(.((((((((((((((((....)))..))))))))))))).)..))))...(((((.......)))))..... ( -29.90, z-score = -1.09, R) >dp4.chr3 11443723 82 - 19779522 -----CUUUUUCGUGUAUAGCGGCUGUCUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGACCGUC -----.......((.(((((((((((((((((....)))..)))))))))))))))).........((((.......))))...... ( -30.60, z-score = -1.20, R) >droAna3.scaffold_13266 15529406 87 + 19884421 UCUUCUCUUCCUCAAUUUAGCGGCUGUUUUGGCUAGCCAGCAAGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC .................(((((((((((((((....)))..)))))))))))).......(((..(((((.......)))))..))) ( -27.40, z-score = -1.25, R) >droEre2.scaffold_4845 8388724 80 - 22589142 -------UUUAUCAUCCUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC -------.(((((((..(((((((((((((((....)))..)))))))))))).....)))))))(((((.......)))))..... ( -29.70, z-score = -1.44, R) >droYak2.chr2R 6125673 82 + 21139217 -----UGUUUAAUCUCUUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC -----............(((((((((((((((....)))..)))))))))))).......(((..(((((.......)))))..))) ( -27.00, z-score = -0.54, R) >droSec1.super_1 11657587 82 - 14215200 -----UGUUUACUCCCAUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC -----...((((..(.((((((((((((((((....)))..))))))))))))).)..))))...(((((.......)))))..... ( -29.90, z-score = -1.09, R) >droSim1.chr2R 12904223 82 - 19596830 -----UGUUUACUCCCAUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC -----...((((..(.((((((((((((((((....)))..))))))))))))).)..))))...(((((.......)))))..... ( -29.90, z-score = -1.09, R) >droVir3.scaffold_12875 497292 80 + 20611582 -----UGCUUGCUUUU--AGCGGCCGUCUUGGCCAGCCAACAAGCGGCUGCAGUAGCUGUGAUAAAUCCAUUUCCGCCGGGACCGUC -----.(((.......--)))(((.(((((((((((((.......)))))(((...)))................)))))))).))) ( -24.30, z-score = 0.27, R) >droMoj3.scaffold_6496 6067191 80 - 26866924 -----UGCUUGCUUUU--AGCGGCCGUUUUGGCCAGCCAACAAGCGGCUGCAGUAGCUGUGAUAAAUCCAUUUCCGCCGGGUCCGAC -----.((..(((...--.(((((((((((((....)))..))))))))))...))).)).....((((.........))))..... ( -24.10, z-score = 0.26, R) >droGri2.scaffold_15245 11421987 80 - 18325388 -----UGCUUGCUUUU--AGCGGCCGUCUUGGCCAGCCAACAAGCGGCUGCUGUUGCUGUGAUAAACCCAUUUCCGCCUGGGCCGUC -----.((..((...(--(((((((((.((((....))))...))))))))))..)).))......((((........))))..... ( -28.40, z-score = -0.75, R) >droPer1.super_4 6847832 82 - 7162766 -----CUUUUUCGUGUAUAGCGGCUGUCUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGACCGUC -----.......((.(((((((((((((((((....)))..)))))))))))))))).........((((.......))))...... ( -30.60, z-score = -1.20, R) >droWil1.scaffold_181141 1667927 87 + 5303230 UGUUUCCGUUUCCAUUGCAGCGGCCGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAAUCCAUUUCCGCCGGGACCGUC ......((.((((..(((((((((((((((((....)))..))))))))))))))((.(.(((......))).).)).)))).)).. ( -32.10, z-score = -1.42, R) >anoGam1.chr2L 8018776 79 - 48795086 --------UUCUCAUUGUUUCAGCCGUACUAGCGACACCCCAGGCGGCGGCCGUCACUGUGCUCAGCCCAUUCCAGACGGGACCGCC --------..............(((.................)))((((((((((...(((.......)))....)))))..))))) ( -19.53, z-score = 0.99, R) >triCas2.ChLG7 12539240 73 + 17478683 -----UGUUUGUUUUUCCAGCGGCAGUGUUGGCAAGCCAGCAAGCAGCAGCAGUGACAGUAAUAAACCCUUAUCCGAC--------- -----.(((((((..(((.((.((..((((((....))))))....)).)).).))....)))))))...........--------- ( -16.40, z-score = -0.05, R) >consensus _____UGUUUACUUUUAUAGCGGCUGUUUUGGCUAGCCAGCAGGCGGCUGCUGUAGCUGUGAUAAACCCGUUUCCGCCGGGUCCGUC ...................(((((((((((((....))))..)))))))))...............(((.........)))...... (-17.72 = -18.19 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:44 2011