| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,165,427 – 14,165,547 |
| Length | 120 |
| Max. P | 0.835100 |

| Location | 14,165,427 – 14,165,547 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.20 |
| Shannon entropy | 0.64331 |
| G+C content | 0.48127 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.74 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
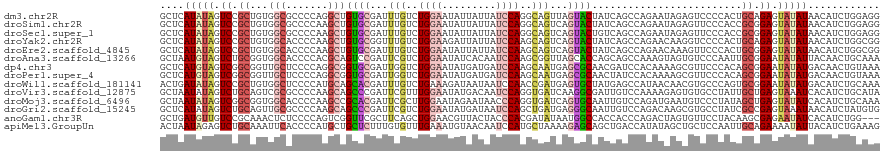
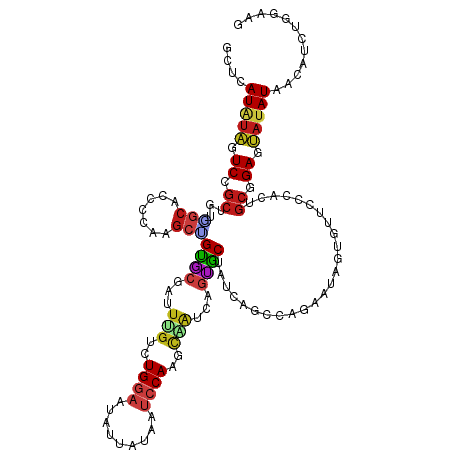
>dm3.chr2R 14165427 120 - 21146708 GCUCAUAUAGUCCGCUGUGGCGCCCCAGGCUGUGCGAUUUGUCUGGAAUAUUAUUAUCCAGGCAGUUAGUACUAUCAGCCAGAAUAGAGUCCCCACUGCAGAGUAUAUAACAUCUGGAGG ((((..((((....))))((((((...))).((((...(((((((((.((....)))))))))))...)))).....)))......)))).....((.((((((.....)).)))).)). ( -37.20, z-score = -1.36, R) >droSim1.chr2R 12902050 120 - 19596830 GCUCAUAUAGUCCGCUGUGGCGCCCCAAGCUGUGCGAUUUGUCUGGAAUAUUAUUAUCCAGGCAGUCAGUACUAUCAGCCAGAAUAGAGUUCCCACCGCGGAGUAUAUAACAUCUGGAGG .((((((((.(((((.((((........(((((((...(((((((((.((....)))))))))))...))))....)))..((((...)))))))).))))).)))))..(....)))). ( -39.00, z-score = -2.22, R) >droSec1.super_1 11655436 120 - 14215200 GCUCAUAUAGUCCGCUGUGGCGCCCCAAGCUGUGCGAUUUGUCUGGAAUAUUAUUAUCCAGGCAGUCAGUACUGUCAGCCAGAAUAGAGUUCCCACCGCGGAGUAUAUAACAUCUGGAGG .((((((((.(((((.((((.((.(....(((.((....((((((((.((....))))))))))(.((....)).).)))))....).))..)))).))))).)))))..(....)))). ( -39.10, z-score = -2.03, R) >droYak2.chr2R 6123527 120 + 21139217 GCUCAUAUAGUCCGCUGUGGCACCCCAAGCUGUGCGGUUUGUCUGGAAGAUUAUUAUCCAAGCAGUCAGUACUAUCAGCCAGAACAAGGUCCCCACUGCAGAGUAUAUAACAUCUGGCGG ....(((((.((.((.((((.(((.((((((....))))))(((((..(((((((..(......)..))))..)))..)))))....)))..)))).)).)).)))))............ ( -29.10, z-score = 0.74, R) >droEre2.scaffold_4845 8386546 120 - 22589142 GCUCAUAUAGUCCGCUGUGGCACCCCAAGCUGUGCGAUUUGUCUGGAAUAUUAUUAUCCAAGCAGUCAGUACUAUCAGCCAGAACAAAGUUCCCACUGCGGAGUAUAUAACAUCUGGCGG ....(((((.(((((.((((........(((((((...(((..((((.((....))))))..)))...))))....)))..((((...)))))))).))))).)))))............ ( -34.30, z-score = -1.57, R) >droAna3.scaffold_13266 15527183 120 + 19884421 GCUAAUGUAGUCUGCGGUGGCACCCCACGCAGUCCGAUUCGUCUGGAAUAUCACAAUCCAAGCGGUUAGCACCAGCAGCCAAAGUAGUGUCCCAAUUGCGGAAUAUAUUACAACUGCAAA ((((((...(.(((((.(((....)))))))).).....((..((((.........))))..))))))))....((((....((((...((((....).)))...))))....))))... ( -34.10, z-score = -1.48, R) >dp4.chr3 11441222 120 - 19779522 GCUCAUGUAGUCGGCGGUUGCUCCCCAGGCGGUGCGAUUGGUCUGGAAUAUGAUGAUCCAAGCAAUGAGCGCAACGAUCCACAAAAGCGUUCCCACAGCGGAAUAUAUGACAACUGUAAA ..(((((((.((.((.((((((..((((((.(......).))))))....((......))))))))((((((..............)))))).....)).)).))))))).......... ( -33.14, z-score = 0.31, R) >droPer1.super_4 6845127 120 - 7162766 GCUCAUGUAGUCGGCGGUUGCUCCCCAGGCGGUGCGAUUGGUCUGGAAUAUGAUGAUCCAAGCAAUGAGCGCAACUAUCCACAAAAGCGUUCCCACAGCGGAAUAUAUGACAACUGUAAA ..(((((((.((.((.((((((..((((((.(......).))))))....((......))))))))((((((..............)))))).....)).)).))))))).......... ( -33.14, z-score = 0.12, R) >droWil1.scaffold_181141 1665239 120 + 5303230 ACUGAUAUAGUCCGCUGUGGCUCCCCAUGCAGCACGAUUUGUCUGAAAGAUAAUAAUCCAACCGAUGAGUGCUAUGAGCCAUAACAACGUGCCCAGUGCGGAAUAUAUGACAUCUGCAAA ....(((((.(((((((((((((.......(((((...(((((.....)))))..(((.....)))..)))))..))))))))...((.......))))))).)))))............ ( -29.80, z-score = -1.16, R) >droVir3.scaffold_12875 494658 120 + 20611582 GCUAAUAUAGUCUGCAGUCGCGCCCCAAGCAGCCCGAUUCGUUUGGAAUAUGACAAUCCAGGUGAUCAAGGCGAUUGUCCAAAAGAGUGUGCCUAUUGCUGAGUAAAUCACAUCUGCAUA .....((((.(((((((((((((.....)).....(((.((..((((.........))))..)))))...)))))))).....))).)))).....(((.((((.....)).)).))).. ( -27.60, z-score = 0.31, R) >droMoj3.scaffold_6496 6064678 120 - 26866924 GCUAAUAUAGUCGGCGGUGGCACCCCAAGCCGCACGAUUCGCUUGGAAUAGAAUAACCCAGGUGAUCAGUGCAAUUGUCCAGAUGAAUGUCCCUAUAGCUGAGUAUAUCACAUCUGCAAA ((..(((((.(((((((.(((.......)))((((...((((((((...........))))))))...))))......)).(((....)))......))))).))))).......))... ( -35.40, z-score = -1.66, R) >droGri2.scaffold_15245 11419248 120 - 18325388 GCUCAUAUAGUCUGCAGUUGCGCCCCAAGCAGCCCGAUUCGUCUGGAAUAUGAUAAUCCAGCUGAUGAGGGCAAUUGUCCAGACAAGCGUGCCUAUCGCCGAGUAAAUAACAUCUAUGUG ((((.....((((((((((((.((.((..((((..((((.(((........)))))))..)))).)).)))))))))..)))))..(((.......))).))))................ ( -35.60, z-score = -1.80, R) >anoGam1.chr3R 5631968 117 - 53272125 GCUGAUGUUGUCCGCAAACUCUCCCCAGUCGGUUCGCUUCAGCUGGAACGUUACUACCCACGAUAUAAUGGCCACCACCCAGACUAGUGUUCCUACAAGCGAGAAUAUCACAUCUGG--- ..(((((((.((.((..(((......))).((..((((....((((..(((........)))......(((...))).))))...))))..)).....)))).))))))).......--- ( -24.70, z-score = 0.17, R) >apiMel3.GroupUn 95136517 120 - 399230636 ACUAAUAGAGUCUGCAAAUUCACCCCAUGCUGCUCUUUGUGUUUGAAAUGUAACAAUCCAUGCUAAAAGAGCAGCUGACCAUAUAGCUGCUCCAAUUGCAGAAAAUAUUACAUCUGAAAG ..(((((...(((((((..............((......((((........))))......)).....(((((((((......)))))))))...)))))))...))))).......... ( -29.90, z-score = -2.89, R) >consensus GCUCAUAUAGUCCGCUGUGGCACCCCAAGCUGUGCGAUUUGUCUGGAAUAUUAUAAUCCAAGCAAUCAGUGCUAUCAGCCAGAAUAGUGUUCCCACUGCGGAGUAUAUAACAUCUGGAAG ....(((((.((.((...(((.......)))((((...(((..((((.........))))..)))...)))).........................)).)).)))))............ (-16.10 = -15.74 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:44 2011