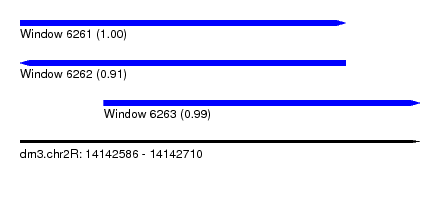
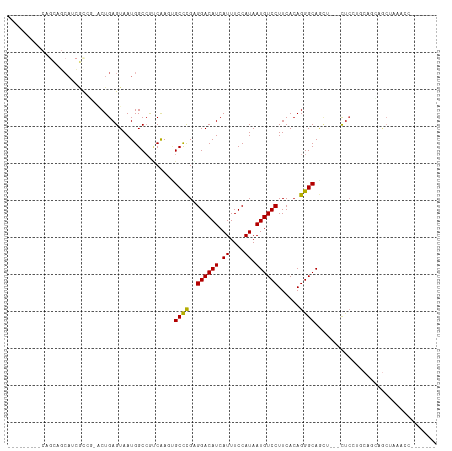
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,142,586 – 14,142,710 |
| Length | 124 |
| Max. P | 0.999398 |

| Location | 14,142,586 – 14,142,687 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.63820 |
| G+C content | 0.55709 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -18.61 |
| Energy contribution | -20.24 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
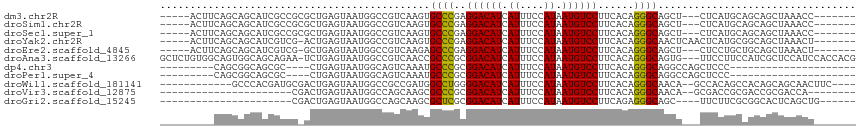
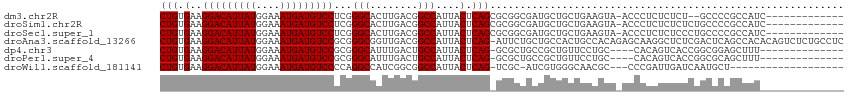
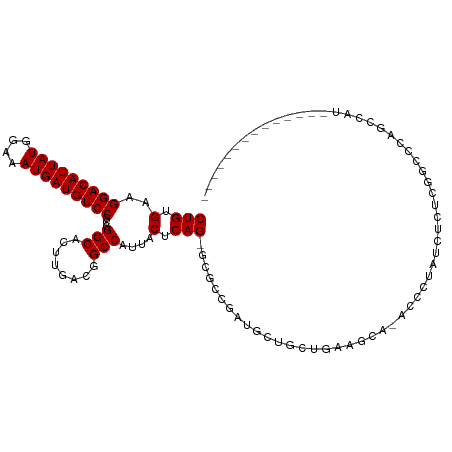
>dm3.chr2R 14142586 101 + 21146708 -------GGUUUAGCUGCUGCAUGAG---AGCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGU----- -------..((((((....((((...---.(((((.(((.(.((((((((((....))))))))))..(((........)))....).))).))))).)))).))))))..----- ( -40.30, z-score = -2.54, R) >droSim1.chr2R 12879299 101 + 19596830 -------GGUUUAGCUGCUGCAUGAG---AGCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGU----- -------..((((((....((((...---.(((((.(((.(.((((((((((....))))))))))..(((........)))....).))).))))).)))).))))))..----- ( -40.30, z-score = -2.54, R) >droSec1.super_1 11632689 101 + 14215200 -------GGUUUAGCUGCUGCAUGAG---AGCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGU----- -------..((((((....((((...---.(((((.(((.(.((((((((((....))))))))))..(((........)))....).))).))))).)))).))))))..----- ( -40.30, z-score = -2.54, R) >droYak2.chr2R 6099613 103 - 21139217 -------AGUUUAGCUGCCGCAUGAGUUGAGUUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGU-CGACGAUGCUGCUGAAGU----- -------..((((((....((((..((..(((.((((...((.(((((((((....))))))))))))))))))..))(((.........))-)....)))).))))))..----- ( -38.70, z-score = -3.08, R) >droEre2.scaffold_4845 8363639 100 + 22589142 -------AGUUUAGCUGCAGCAGGAG---AGCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCUCUUGACGGCCAUUACUCAGC-CGACGAUGCUGCUGAAGU----- -------......(((.(((((((((---((((......(((.(((((((((....)))))))))))))))))))..((((.........))-))......)))))).)))----- ( -39.80, z-score = -2.78, R) >droAna3.scaffold_13266 15502618 112 - 19884421 CGUGGUGGAUGGAGCGAUGGAAGGAA---CACUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCGGUUGACGGCCAUUACUCAGA-UUCUGCUGCCACUGCCACAGAGC .((((..(.(((((..((((..(..(---.(((((((.((...(((((((((....)))))))))))))))))))..)..))))..))(((.-.....)))))))..))))..... ( -48.00, z-score = -3.99, R) >dp4.chr3 11417770 82 + 19779522 ---------------------GGGAGCUGGCCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCAUUUGACUGCCAUUACUCAG----GCGCUGCCGCUG--------- ---------------------...(((.(((..((((((.(..(((((((((....)))))))))...((((......))))....).)))----).)).)))))).--------- ( -36.90, z-score = -3.39, R) >droPer1.super_4 6821556 82 + 7162766 ---------------------GGGAGCUGGCCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCAUUUGACUGCCAUUACUCAG----GCGCUGCCGCUG--------- ---------------------...(((.(((..((((((.(..(((((((((....)))))))))...((((......))))....).)))----).)).)))))).--------- ( -36.90, z-score = -3.39, R) >droWil1.scaffold_181141 1629602 98 - 5303230 ----GAAGUUGCUGCUGUGGCUGUGGC--UGUUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCCCAGGCCAUCGGCGGCCAUUACUCAGUCGCAUCGUGGGC------------ ----...(.(((.((((.(...(((((--(((((.((((.(..(((((((((....))))))))))))))....))))))))))..).)))).))).)......------------ ( -41.90, z-score = -2.44, R) >droVir3.scaffold_12875 463264 84 - 20611582 --------UGGUCGCGGUCGCGGUCGC--UGUUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCGCUUGCUGGCCAUUACUCAGUCG---------------------- --------(((((((((..(((.((((--..(..(...)..).(((((((((....))))))))))))).))))))).)))))...........---------------------- ( -33.50, z-score = -1.44, R) >droGri2.scaffold_15245 11391396 84 + 18325388 ------CAGCUGAGUGCCGCGAAGAA----GCUGCCCUCUGAAGGACAUUAUGGAAAUGAUGUCCGCGAGCGCUUGCUGGCCAUUACUCAGUCG---------------------- ------((((.(((((((((..(((.----.......)))...(((((((((....)))))))))))).))))))))))...............---------------------- ( -33.50, z-score = -2.99, R) >consensus _______GGUUUAGCUGCGGCAGGAG___AGCUGCCCUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCACUUGACGGCCAUUACUCAGC_CGGCGAUGCUGCUG_________ ..............................((((....((((.(((((((((....)))))))))...(((........))).)))).))))........................ (-18.61 = -20.24 + 1.62)

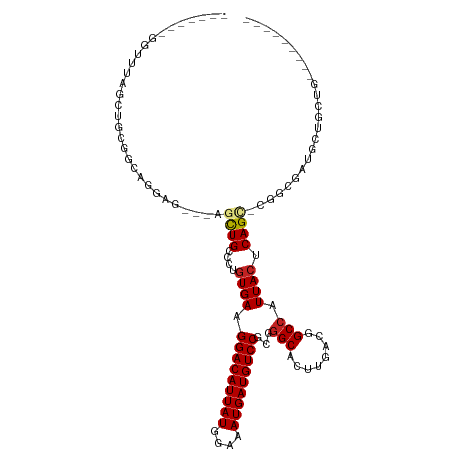
| Location | 14,142,586 – 14,142,687 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.63820 |
| G+C content | 0.55709 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.15 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14142586 101 - 21146708 -----ACUUCAGCAGCAUCGCCGCGCUGAGUAAUGGCCGUCAAGUGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGCU---CUCAUGCAGCAGCUAAACC------- -----..((((((.((......))))))))...((((.((....(((((((((((((.((....)).))))))))....)))))((.---.....)).)).))))....------- ( -32.20, z-score = -1.90, R) >droSim1.chr2R 12879299 101 - 19596830 -----ACUUCAGCAGCAUCGCCGCGCUGAGUAAUGGCCGUCAAGUGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGCU---CUCAUGCAGCAGCUAAACC------- -----..((((((.((......))))))))...((((.((....(((((((((((((.((....)).))))))))....)))))((.---.....)).)).))))....------- ( -32.20, z-score = -1.90, R) >droSec1.super_1 11632689 101 - 14215200 -----ACUUCAGCAGCAUCGCCGCGCUGAGUAAUGGCCGUCAAGUGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGCU---CUCAUGCAGCAGCUAAACC------- -----..((((((.((......))))))))...((((.((....(((((((((((((.((....)).))))))))....)))))((.---.....)).)).))))....------- ( -32.20, z-score = -1.90, R) >droYak2.chr2R 6099613 103 + 21139217 -----ACUUCAGCAGCAUCGUCG-ACUGAGUAAUGGCCGUCAAGUGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAACUCAACUCAUGCGGCAGCUAAACU------- -----........(((...((((-..(((((..((.....))..(((((((((((((.((....)).))))))))....))))).....)))))..)))).))).....------- ( -28.20, z-score = -1.38, R) >droEre2.scaffold_4845 8363639 100 - 22589142 -----ACUUCAGCAGCAUCGUCG-GCUGAGUAAUGGCCGUCAAGAGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGCU---CUCCUGCUGCAGCUAAACU------- -----......((((((..(.((-(((.......))))).)....((((((((((((.((....)).))))))))....))))....---....)))))).........------- ( -35.50, z-score = -2.67, R) >droAna3.scaffold_13266 15502618 112 + 19884421 GCUCUGUGGCAGUGGCAGCAGAA-UCUGAGUAAUGGCCGUCAACCGCCCGCGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGUG---UUCCUUCCAUCGCUCCAUCCACCACG ..(((((.((....)).))))).-...(((..((((......((.((((..((((((.((....)).))))))......)))).)).---......))))..)))........... ( -31.62, z-score = -0.83, R) >dp4.chr3 11417770 82 - 19779522 ---------CAGCGGCAGCGC----CUGAGUAAUGGCAGUCAAAUGCCCGCGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGGCCAGCUCCC--------------------- ---------.((((((.((.(----(((.((...((((......)))).))((((((.((....)).))))))....))))))..))).)))...--------------------- ( -28.50, z-score = -1.64, R) >droPer1.super_4 6821556 82 - 7162766 ---------CAGCGGCAGCGC----CUGAGUAAUGGCAGUCAAAUGCCCGCGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGGCCAGCUCCC--------------------- ---------.((((((.((.(----(((.((...((((......)))).))((((((.((....)).))))))....))))))..))).)))...--------------------- ( -28.50, z-score = -1.64, R) >droWil1.scaffold_181141 1629602 98 + 5303230 ------------GCCCACGAUGCGACUGAGUAAUGGCCGCCGAUGGCCUGGGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAACA--GCCACAGCCACAGCAGCAACUUC---- ------------........(((..(((.....((((.((.(.((.(((((((((((.((....)).)))))))...)))).)).).--))....)))))))..))).....---- ( -28.70, z-score = -0.43, R) >droVir3.scaffold_12875 463264 84 + 20611582 ----------------------CGACUGAGUAAUGGCCAGCAAGCGCCCGCGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAACA--GCGACCGCGACCGCGACCA-------- ----------------------((.(((.......((......))((((..((((((.((....)).))))))......))))..))--)))..(((....)))....-------- ( -22.40, z-score = -0.21, R) >droGri2.scaffold_15245 11391396 84 - 18325388 ----------------------CGACUGAGUAAUGGCCAGCAAGCGCUCGCGGACAUCAUUUCCAUAAUGUCCUUCAGAGGGCAGC----UUCUUCGCGGCACUCAGCUG------ ----------------------.(.((((((....(((.....((.(((..((((((.((....)).))))))....))).)).((----......))))))))))))..------ ( -27.90, z-score = -1.37, R) >consensus _________CAGCAGCAUCGCCG_ACUGAGUAAUGGCCGUCAAGUGCCCGAGGACAUCAUUUCCAUAAUGUCCUUCACAGGGCAGCU___CUCCUGCAGCAGCUAAACC_______ .............................................((((..((((((.((....)).))))))......))))................................. (-13.32 = -13.15 + -0.16)

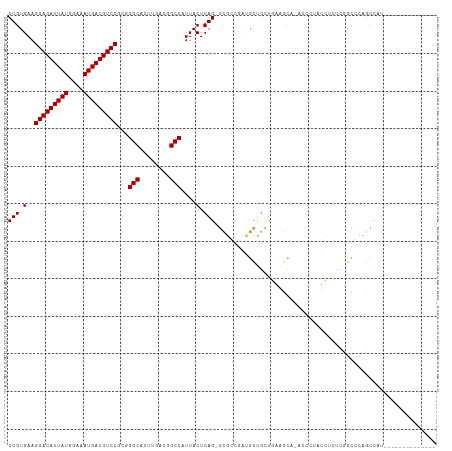
| Location | 14,142,612 – 14,142,710 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.53334 |
| G+C content | 0.55843 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -18.14 |
| Energy contribution | -19.64 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14142612 98 + 21146708 CUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGUA-ACCCUCUCUCU--GCCCCGCCAUC------------- ..(((.((((((((((....)))))))))).(((((...((.((...((((((((((((....))).)))).))))-))).))....)--)))))))....------------- ( -38.10, z-score = -3.53, R) >droSim1.chr2R 12879325 100 + 19596830 CUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGUA-ACCCUCUCUCUCUGCCCCGCCAUC------------- ..(((.((((((((((....)))))))))).(((((...((.((...((((((((((((....))).)))).))))-)....)).))..))))))))....------------- ( -37.00, z-score = -3.09, R) >droSec1.super_1 11632715 100 + 14215200 CUGUGAAGGACAUUAUGGAAAUGAUGUCCUCGGGCACUUGACGGCCAUUACUCAGCGCGGCGAUGCUGCUGAAGUA-ACCCUCUCUCCCUGCCCCGCCAUC------------- ..(((.((((((((((....)))))))))).(((((...((.((...((((((((((((....))).)))).))))-)..)).))....))))))))....------------- ( -37.00, z-score = -3.08, R) >droAna3.scaffold_13266 15502651 113 - 19884421 CUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCGGUUGACGGCCAUUACUCAG-AUUCUGCUGCCACUGCCACAGAGCAAGGCUCUCGACUCAGCCACACAGUCUCUGCCUC (((((..(((((((((....)))))))))...(((((((((.((((......(((-......)))....(((......))).)))).))))))..))).))))).......... ( -40.80, z-score = -1.82, R) >dp4.chr3 11417785 95 + 19779522 CUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCAUUUGACUGCCAUUACUCAG-GCGCUGCCGCUGUUCCUGC----CACAGUCACCGGCGGAGCUUU-------------- ..((((.(((((((((....)))))))))...((((......)))).))))..((-((.((((((.((...(((.----..))).)).)))))).)))).-------------- ( -37.00, z-score = -2.21, R) >droPer1.super_4 6821571 95 + 7162766 CUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCAUUUGACUGCCAUUACUCAG-GCGCUGCCGCUGUUCCUGC----CACAGUCACCGGCGCAGCUUU-------------- (((.(..(((((((((....)))))))))...((((......))))....).)))-((((((..(((((......----.)))))...))))))......-------------- ( -36.40, z-score = -1.98, R) >droWil1.scaffold_181141 1629632 90 - 5303230 CUGUGAAGGACAUUAUGGAAAUGAUGUCCCCAGGCCAUCGGCGGCCAUUACUCAG-UCGC-AUCGUGGGCAACGC---CCCGAUUGAUCAAUGCU------------------- ..((((.(((((((((....)))))))))...((((......)))).))))...(-(((.-((((.((((...))---)))))))))).......------------------- ( -34.80, z-score = -2.27, R) >consensus CUGUGAAGGACAUUAUGGAAAUGAUGUCCGCGGGCACUUGACGGCCAUUACUCAG_GCGCCGAUGCUGCUGAAGCA_ACCCUAUCUCUCGGCCCAGCCAU______________ ..((((.(((((((((....)))))))))...(((........))).))))............................................................... (-18.14 = -19.64 + 1.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:42 2011