| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,131,662 – 14,131,740 |
| Length | 78 |
| Max. P | 0.914800 |

| Location | 14,131,662 – 14,131,740 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.56764 |
| G+C content | 0.44620 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -10.79 |
| Energy contribution | -10.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
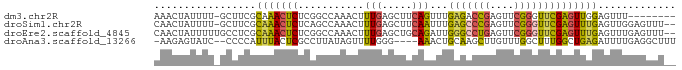
>dm3.chr2R 14131662 78 + 21146708 AAACUAUUUU-GCUUCGCAAACUCUCGGCCAAACUUUGAGCUUCAGUUUGAGACCGAGUUCGGGUUCGAGUUGGAGUUU-------- .......(((-((...)))))................((((((((..(((((.(((....))).)))))..))))))))-------- ( -19.80, z-score = -0.22, R) >droSim1.chr2R 12867240 84 + 19596830 CAACUAUUUU-GCUUCGCAAACUCUCAGCCAAACUUUGAGCUUCAAUUUGAGCCCGAGUUCGGGUUCGAGUUUGAGUUGGAGUUU-- ..........-.......((((((.(((((((((.(((.....))).(((((((((....)))))))))))))).))))))))))-- ( -29.40, z-score = -3.33, R) >droEre2.scaffold_4845 8352768 85 + 22589142 CAACUAUUUUUGCCUCGCAAACUCUCGGCCAAACUUUGAGCUGCAGAUUGGGCCUGAGUUCGGGUUCGAGUUUGAGUUUGAGUUU-- .............(((((((((((.((((((.....)).))))......(((((((....))))))))))))))....))))...-- ( -23.40, z-score = -0.38, R) >droAna3.scaffold_13266 15490398 80 - 19884421 -AAGAGUAUC--CCCCAUUUACUCGCCUUAUAGUUUUGGG----AAACUGCAAGCUUGUUUGGCUUUGGCUGAGAUUUUGAGGCUUU -..(((((..--.......)))))((((((.((((((((.----...))((.((((.....))))...)).)))))).))))))... ( -24.10, z-score = -1.79, R) >consensus CAACUAUUUU_GCCUCGCAAACUCUCGGCCAAACUUUGAGCUUCAAAUUGAGACCGAGUUCGGGUUCGAGUUUGAGUUUGAGUUU__ .................(((((((...........(((.....)))...(((((((....))))))))))))))............. (-10.79 = -10.85 + 0.06)
| Location | 14,131,662 – 14,131,740 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.56764 |
| G+C content | 0.44620 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
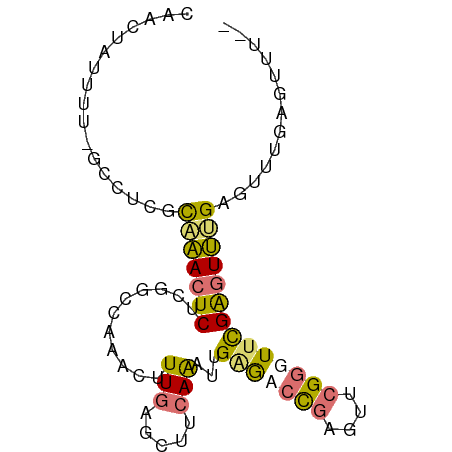
>dm3.chr2R 14131662 78 - 21146708 --------AAACUCCAACUCGAACCCGAACUCGGUCUCAAACUGAAGCUCAAAGUUUGGCCGAGAGUUUGCGAAGC-AAAAUAGUUU --------(((((.....(((....))).((((((..((((((.........))))))))))))..(((((...))-)))..))))) ( -17.80, z-score = -0.88, R) >droSim1.chr2R 12867240 84 - 19596830 --AAACUCCAACUCAAACUCGAACCCGAACUCGGGCUCAAAUUGAAGCUCAAAGUUUGGCUGAGAGUUUGCGAAGC-AAAAUAGUUG --......((((((((((((..(.(((((((.((((((.....).)))))..))))))).)..)))))))......-.....))))) ( -23.60, z-score = -2.36, R) >droEre2.scaffold_4845 8352768 85 - 22589142 --AAACUCAAACUCAAACUCGAACCCGAACUCAGGCCCAAUCUGCAGCUCAAAGUUUGGCCGAGAGUUUGCGAGGCAAAAAUAGUUG --.((((..........((((....(((((((.((((.((..(........)..)).))))..)))))))))))........)))). ( -19.97, z-score = -0.99, R) >droAna3.scaffold_13266 15490398 80 + 19884421 AAAGCCUCAAAAUCUCAGCCAAAGCCAAACAAGCUUGCAGUUU----CCCAAAACUAUAAGGCGAGUAAAUGGGG--GAUACUCUU- ....(((((....(((.(((.((((.......))))..(((((----....)))))....))))))....)))))--.........- ( -18.50, z-score = -1.25, R) >consensus __AAACUCAAACUCAAACUCGAACCCGAACUCGGGCUCAAACUGAAGCUCAAAGUUUGGCCGAGAGUUUGCGAAGC_AAAAUAGUUG ..................(((....(((((((.((((.(((((........))))).))))..)))))))))).............. (-10.74 = -10.42 + -0.31)


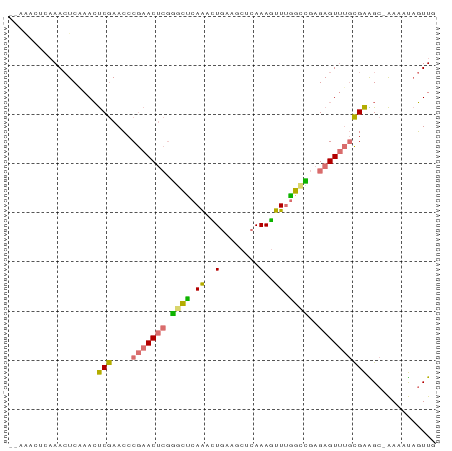
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:40 2011