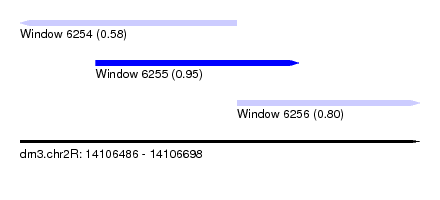
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,106,486 – 14,106,698 |
| Length | 212 |
| Max. P | 0.951856 |

| Location | 14,106,486 – 14,106,601 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Shannon entropy | 0.40857 |
| G+C content | 0.53648 |
| Mean single sequence MFE | -38.71 |
| Consensus MFE | -25.95 |
| Energy contribution | -25.11 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14106486 115 - 21146708 -AGGGGUAUUGGGGGCAGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCAGCGAUUUAUGCUUAAAACGGUGCAGAUUCCUGUGCCGCAGC -.............((.((.((((((((((.((((.((((((....))))))((.......))...)))).)(((.((.((((....)))).)).))).))))))))).))))... ( -42.30, z-score = -1.12, R) >droPer1.super_4 6785849 116 - 7162766 AGGCAGAGCACAAGCCGGAUCAGAGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCAGCGGCGAUUUGUGCUCAAAACGGUGUAGAUUUGUAUGCCACAGC .((((((((((....(......).(((((((.((..((((((....)))))).......((((....)))).)).)))))))))))))...((((.......)))).))))..... ( -41.10, z-score = -1.71, R) >dp4.chr3 11382525 116 - 19779522 AGGCAGAGCACAAGCCGGAUCAGAGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCAGCGGCGAUUUGUGCUUAAAACGGUGUAGAUUUGUAUGCCACAGC .((((..(((...((((....((.(((((((.((..((((((....)))))).......((((....)))).)).)))))))...)).....)))).......))).))))..... ( -39.30, z-score = -1.36, R) >droAna3.scaffold_13266 15461741 115 + 19884421 -GGGGUUCCUGGAGGCAGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCUGCGGCGAUUUAUGCUUAAAACGGUGUAGAUUCUGGUGCCGCAAA -...(((((((....))))))).......((((((.((((((....)))))).......((((....))))..(((.((((((((((......))))))))))))).))))))... ( -40.80, z-score = -0.83, R) >droEre2.scaffold_4845 8327111 114 - 22589142 -AGGGGAAUU-GGGGCUGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCAGCGAUUUAUGCUUAAAACGGUGCAGAUUCCUGUGCCCCAGC -.......((-(((((....((((((((((.((((.((((((....))))))((.......))...)))).)(((.((.((((....)))).)).))).)))))))))))))))). ( -46.50, z-score = -2.15, R) >droYak2.chr2R 6062442 114 + 21139217 -AGGGGAAUU-GGGGCUGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCAGCGAUUUAUGCUUAAAACGGUGCAGAUUCCUGUGCCGCAGC -.........-...((((((((((((((((.((((.((((((....))))))((.......))...)))).)(((.((.((((....)))).)).))).))))))))).)).)))) ( -43.30, z-score = -1.39, R) >droSec1.super_1 11596901 115 - 14215200 -AGGGGGAUUGGGGGCAGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCAGCGAUUUAUUCUUAAAACGGUGCAGAUUCCUGUGCCGCAGC -.............((.((.((((((((((.((((.((((((....))))))((.......))...)))).)(((.((.((((....)))).)).))).))))))))).))))... ( -42.30, z-score = -1.37, R) >droSim1.chr2R 12842246 115 - 19596830 -AGGGGGAUUGGGGGCAGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCAGCGAUUUAUGCUUAAAACGGUGCAGAUUCCUGUGCCGCAGC -.............((.((.((((((((((.((((.((((((....))))))((.......))...)))).)(((.((.((((....)))).)).))).))))))))).))))... ( -42.30, z-score = -1.03, R) >droVir3.scaffold_12875 418101 104 + 20611582 -----------AGUCAAGAAACGAGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUC-GGCGCAUGCCUCGCGGCGAUUUAUGCUUAAAACGGUGUAGAUUUUUAUGCUGUUGC -----------.((((....((....))..))))..((((((....))))))(((((((-(.(((.......))).)))..)))))....((((((((((....)))))))))).. ( -31.60, z-score = -0.88, R) >droGri2.scaffold_15245 11346402 101 - 18325388 -----------AGUCAAGAAACGAGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUC-GGCGCAUGCCUGUCGGCGAUUUAUGCUUAAAACGGUGCAGAUUCUUAUGUUGC--- -----------...........(((((((...(((.((((((....))))))(((((((-(.((((....)).)).)))..))))).........))).))))))).......--- ( -27.80, z-score = -0.05, R) >droMoj3.scaffold_6496 5978831 100 - 26866924 ----------GAGACUUG-ACUUUGAAUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUC-GGCGCAUGCCUUGCGGCGAUUUAUGCUUAAAACGGUGUAGAUUUUUAUGCUG---- ----------.....(((-(...((((((((.((((((((((....)))))).......-(((....))))))).))))))))...))))..((((((((....))))))))---- ( -28.50, z-score = -0.98, R) >consensus _AGGGGAAUUGGGGGCAGGGACAGGAGUCGUGGCAAGUUCAGUGAGUUGAACGCAUAUCGGGCGCAUGCCCCGCGGCGAUUUAUGCUUAAAACGGUGCAGAUUCCUGUGCCGCAGC ......................(((((((..((((.((((((....))))))((.......))...))))..(((.((.((((....)))).)).))).))))))).......... (-25.95 = -25.11 + -0.84)


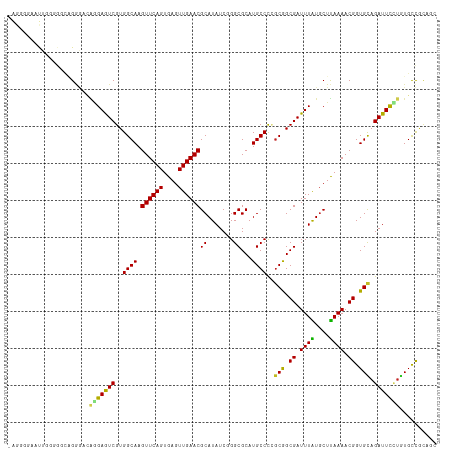
| Location | 14,106,526 – 14,106,634 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Shannon entropy | 0.38642 |
| G+C content | 0.61872 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14106526 108 + 21146708 CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCUGCCCCCAAUACCCCUGUUCACCCCACACCUACCCCCCCUCAUUGCCGA .((.((((((.((((.......))(((((......)))))..))...(((....)))..))))))))......................................... ( -20.90, z-score = -1.42, R) >droAna3.scaffold_13266 15461781 92 - 19884421 CCGCAGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCUGCCUCCAGGAACCCCCUACCCUCCUUGCCGAU---------------- ..(((((((((((....).)))))(((((......))))).......(((....))))))))....(((.....)))...............---------------- ( -21.10, z-score = -1.26, R) >droEre2.scaffold_4845 8327151 92 + 22589142 CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCAGCCCCAAUUCCCCUAUUCCCCCUCAUUGCCGA---------------- ....(((((..((((.......))(((((......)))))..))...(((....)))...)))))...........................---------------- ( -20.50, z-score = -1.23, R) >droYak2.chr2R 6062482 95 - 21139217 CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCAGCCCCAAUUCCCCUCUACCCCCCCUCCGUUGCCGA------------- ....(((((..((((.......))(((((......)))))..))...(((....)))...)))))..............................------------- ( -20.50, z-score = -1.06, R) >droSec1.super_1 11596941 107 + 14215200 CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCUGCCCCCAAUCCCCCUGUUCACCCCACUCCUACCCCCC-UCAUUCCCGA .((.((((((.((((.......))(((((......)))))..))...(((....)))..))))))))..............................-.......... ( -20.90, z-score = -2.07, R) >droSim1.chr2R 12842286 107 + 19596830 CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCUGCCCCCAAUCCCCCUGUUCACCCCACUCCUACCCCCC-UCAUUGUCGA .((.((((((.((((.......))(((((......)))))..))...(((....)))..))))))))..............................-.......... ( -20.90, z-score = -1.41, R) >consensus CUGCGGGGCAUGCGCCCGAUAUGCGUUCAACUCACUGAACUUGCCACGACUCCUGUCCCUGCCCCCAAUCCCCCUGUUCACCCCACUCCUACCCC_____________ ....(((((..((((.......))(((((......)))))..))...(((....)))...)))))........................................... (-18.72 = -18.75 + 0.03)

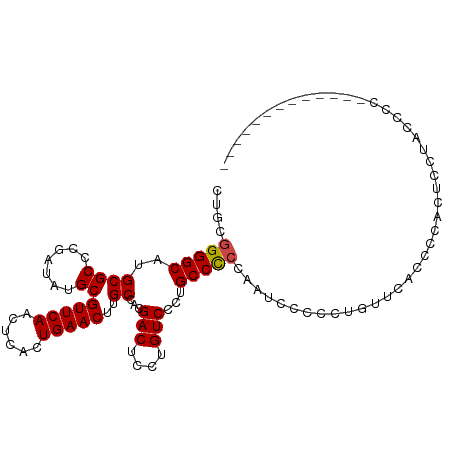

| Location | 14,106,601 – 14,106,698 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Shannon entropy | 0.29383 |
| G+C content | 0.59697 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14106601 97 + 21146708 GUUCACCCCACACCUACCCCCCCUC-AUUGCCGACCGGCUUCUGAGGCUCCUGCUCCCGCUUCCUUUGGC-------UGGAGCUGAAUCAGCUCCACCUACUUCA .....................((((-(..(((....)))...))))).....((....)).......((.-------((((((((...))))))))))....... ( -26.50, z-score = -3.17, R) >droEre2.scaffold_4845 8327225 81 + 22589142 ---------------AUUCCCCCUC-AUUGCCGACCGGCUUCUGAUGCUCCUGCUCCCGCUUCCUUGGUU--------GGAGCUGAAUCAGCUCCACCUACUUCA ---------------........((-(..(((....)))...))).((....))............(((.--------(((((((...))))))))))....... ( -21.20, z-score = -2.44, R) >droYak2.chr2R 6062556 92 - 21139217 -------------CUACCCCCCCUCCGUUGCCGACCGGCUUCUGAUGCUCCUGCUCCCGCUUCCUUGGUUCCUUGGCUGGAGCUGAAUCAGCUCCACCUACUUCA -------------.............(..(((((..(((....((.((....))))..)))...)))))..)..((.((((((((...))))))))))....... ( -24.00, z-score = -2.05, R) >droSec1.super_1 11597016 95 + 14215200 GUUCACCCCACUCCUACCCCCC-UC-AUUCCCGACCGGCUUCUGAGGCUCCUGCUCCCGCUUCCUU-GGC-------UGGAGCUGAAUCAGCUCCACCUACUUCA ....................((-((-(...((....))....))))).....((....))......-((.-------((((((((...))))))))))....... ( -22.00, z-score = -1.66, R) >droSim1.chr2R 12842361 95 + 19596830 GUUCACCCCACUCCUACCCCCC-UC-AUUGUCGACCGGCUUCUGAGGCUCCUGCUCCCGCUUCCUU-GGC-------UGAAGCUGAAUCAGCUCCACCUACUUCA ....................((-((-(..(((....)))...))))).....((....))......-((.-------((.(((((...))))).))))....... ( -17.00, z-score = -0.20, R) >consensus GUUCACCCCAC_CCUACCCCCCCUC_AUUGCCGACCGGCUUCUGAGGCUCCUGCUCCCGCUUCCUU_GGC_______UGGAGCUGAAUCAGCUCCACCUACUUCA .............................(((....)))....(((((..........)))))...............(((((((...))))))).......... (-15.48 = -16.12 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:36 2011