
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,081,803 – 14,081,927 |
| Length | 124 |
| Max. P | 0.634567 |

| Location | 14,081,803 – 14,081,927 |
|---|---|
| Length | 124 |
| Sequences | 7 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.58582 |
| G+C content | 0.58150 |
| Mean single sequence MFE | -49.23 |
| Consensus MFE | -14.73 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14081803 124 + 21146708 AUUGCCCGUUCCCAUUCCGGUUGACAAAGUC-GCCGAGG---AGCUGGGGAUCUGUGGCC-UCAACGU----AUUCUUGGCGAUUCGGGGAGC---UACG-GCACUAAAGAGAUGGAGAUGGUCGAGGUGUAACGGU .(((.((((..((((..(...........((-.(((((.---.((..(((((.(((....-...))).----)))))..))..))))).))((---....-))......)..))))..)))).)))........... ( -42.60, z-score = -0.11, R) >droPer1.super_4 6761291 101 + 7162766 AUUGCCCAUUUCCAUUCCGGUUGC----GCC-GCUGAUG--AAGACGCGCUGCUGCGG-------------------CAGAGGUAAGCAGCGG---UGCG-GUGGGG-----AUG-AGAUGGGCGAGGUGUAACGGU .(((((((((((.(((((.(..((----(((-((((...--...((.(.((((....)-------------------))).)))...))))))---))).-.).)))-----)))-))))))))))........... ( -52.70, z-score = -4.53, R) >dp4.chr3 11358050 106 + 19779522 AUUGCCCAUUUCCAUUCCGGUUGC----GCC-GCUGAUG--AAGACGCGCUGCUGCGG-------------------CAGAGGUAAGCAGCGG---UGCG-GUGCGGUGCGGAUG-AGAUGGGCGAGGUGUAACGGU .(((((((((((...(((....((----(((-((.....--....(((((((((((.(-------------------(....))..)))))))---))))-..)))))))))).)-))))))))))........... ( -52.80, z-score = -3.25, R) >droAna3.scaffold_13266 15433370 113 - 19884421 AUUGCCCAUUUCCAUUCCGGUUGACAAAGUU--AUGUUG----GCCGGGGGGCAUUGG----------------UCUUGGGGACUGGGGGGCUAGAUCCGAGGAUCGUAGA--UGGAGAUGGGCGAGGUGUAACGGU .((((((((((((((((((((..(((.....--.)))..----)))))........((----------------((((.(((.((((....)))).))).))))))...))--)))))))))))))........... ( -54.30, z-score = -5.36, R) >droYak2.chr2R 6036165 134 - 21139217 AUUGCCCGUUUCCAUUCCGGUUGACAAAGUCAGCCGAGGCGGAGUUGGGGAUCUCUGGCCACCGAGAUGGAGAUUCGGGGAGAUUCGGGGUGC---UCCGUGUGCCGCAGAGAUGGAGAUGGUCGAGGUGUAACGGU .(((.(((((((((((.((((((((...))))))))..((((...(((((..((((((((.(((((.......)))))))....))))))..)---))))....))))...))))))))))).)))........... ( -54.10, z-score = -1.39, R) >droSec1.super_1 11572107 128 + 14215200 AUUGCCCGUUCCCAUUCCGGUUGACAAAGUC-GCCGAGG---AGCUGGGGAUCUGUGGCCUCAAAGAU----UUCUUUGGCGAUUCGGGCACCGAAUCCG-GCACCGAAGAGAUGGAGAUGGUCGAGGUGUAACGGU .((((((((((((((((((((.(((...)))-)))..))---)).)))))))...(((((((....((----((((((((.(((((((...)))))))..-...))))))))))...)).))))).)).)))).... ( -44.70, z-score = 0.08, R) >droSim1.chr2R 12815983 125 + 19596830 AUUGCCCGUUCCCAUUCCGGUUGACAAAGUC-GCCGAGG---AGCUGGGGAUCUGUGGCCACAAAGAU----UUCUUUGGCGAUUCGGGCAGC---UCCG-GCACCGAAGAGAUGGAGAUGGUCGAGGUGUAACGGU .(((.((((..(((((.((((.(((...)))-(((..((---(((((.(((((....((((..(((..----..))))))))))))...))))---))))-))))))....)))))..)))).)))........... ( -43.40, z-score = 0.14, R) >consensus AUUGCCCGUUUCCAUUCCGGUUGACAAAGUC_GCCGAGG___AGCUGGGGAUCUGUGGCC____A__U_____UUCUUGGAGAUUCGGGGAGC___UCCG_GCACCGAAGAGAUGGAGAUGGUCGAGGUGUAACGGU .(((.(((((.(((((.((((.((.....)).)))).........((((..(((((..............................))))).....))))...........))))).))))).)))........... (-14.73 = -15.32 + 0.60)
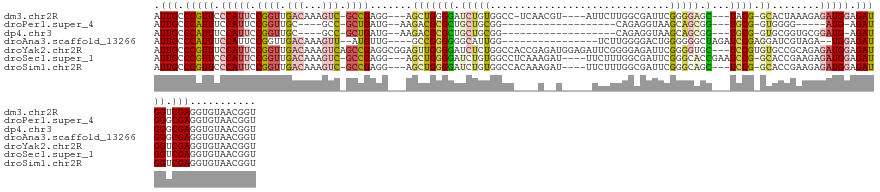

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:34 2011