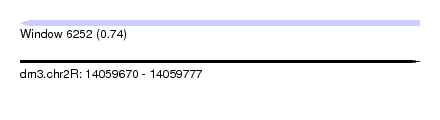
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,059,670 – 14,059,777 |
| Length | 107 |
| Max. P | 0.744663 |

| Location | 14,059,670 – 14,059,777 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.82071 |
| G+C content | 0.30751 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -2.85 |
| Energy contribution | -3.33 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.12 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14059670 107 - 21146708 CCAAUGGAAACAAGUUAAUGUGCGCAUAAUAUGCAAA-UUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAUACAUACAUAU-GUACGCAAGUACAUAGUACAUAU ....((....)).....((((((((((...))))...-.....((((((((((((....)))))..))))))).........(((-((((....))))))))))))).. ( -32.40, z-score = -4.31, R) >droSim1.chr2R 12792200 107 - 19596830 CCAAUGGAAACAAGUUAAUGUGCGCAUAAUAUGCAAA-UUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAGACAUACAUAU-GUUCGAAAGUACAUAGUACAUAU ....((....)).....((((((((((...))))...-...((((((((((((((....)))))..)))))).)))......(((-((.(....).))))))))))).. ( -28.30, z-score = -3.34, R) >droSec1.super_1 11549483 98 - 14215200 CCAAUGAAAACAAGUUAAUGUGCGCAUAAUAUGCAAA-UUUUCUGUUUGUUUUGGUUUGCCAAAAUACAA---------ACAUAU-GUACGCAAGUACAUAGUACAUAU .................((((((((((...))))...-.....((((((((((((....)))))..))))---------)))(((-((((....))))))))))))).. ( -29.70, z-score = -4.23, R) >droYak2.chr2R 6012733 107 + 21139217 UCAAUAGAAUCAAGGAAAUAUGCACAUAAUAUGCAAA-UUUUCUGUUUGUUUUGGUUCGCCAAGAUACAAACAAUACAUACAUAC-GUAAGCAAUUACAUGGUACAUAC ......(.(((((((((((.((((.......)))).)-))))))(((((((((((....)))))..)))))).............-((((....)))).)))).).... ( -22.90, z-score = -2.87, R) >droAna3.scaffold_13266 15409691 96 + 19884421 CUGGCUAAAAUACUUCAAAUUCCACCUGAAAUAAAGC-UUUCGUAUUUGUUUUGGUUUGCCUGCAUACAUAUAAGAAGUAUAUAU-GUACCUGCGUAC----------- ..(((..................(((.((((((((((-....)).)))))))))))..))).(((((((((((.......)))))-)))..)))....----------- ( -15.35, z-score = -0.13, R) >dp4.chr2 15323744 102 + 30794189 ---UCCUUUAAAACUUAAAGCAUAUAUAGUUUCAA---UUUUCAAUUAAUUCUGUUUUACUCUAGUAUCAGCAGUAUAUACAUAC-AUAUAUGUGUACACUGUACAUAC ---............(((((((....(((((....---.....)))))....))))))).....((((..(((((.((((((((.-...)))))))).)))))..)))) ( -16.20, z-score = -1.79, R) >droGri2.scaffold_15245 5679673 93 + 18325388 ---ACAAUAUUAAAAAGCUUUGCCUGGAAUUUGUUGUGUUUUGGAUUUGUCAAAAUAUG-CAAAACACACACAA-ACACACACAUAAUACAUGUGUGC----------- ---..........................((((((((((((((.((.(......).)).-))))))))).))))-)..(((((((.....))))))).----------- ( -20.90, z-score = -1.07, R) >consensus CCAAUGAAAACAAGUUAAUGUGCACAUAAUAUGCAAA_UUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAUACAUACAUAU_GUACGCAAGUACAUAGUACAUAU ...............................................(((((.((....)).)))))...................((((....))))........... ( -2.85 = -3.33 + 0.47)

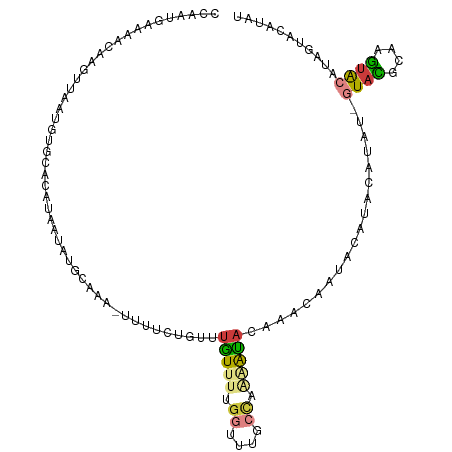

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:33 2011