| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,030,682 – 14,030,826 |
| Length | 144 |
| Max. P | 0.975135 |

| Location | 14,030,682 – 14,030,826 |
|---|---|
| Length | 144 |
| Sequences | 12 |
| Columns | 178 |
| Reading direction | forward |
| Mean pairwise identity | 72.10 |
| Shannon entropy | 0.53658 |
| G+C content | 0.40918 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -16.57 |
| Energy contribution | -15.68 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
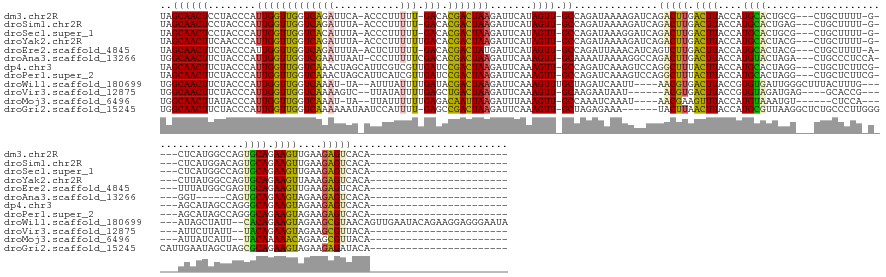
>dm3.chr2R 14030682 144 + 21146708 UAGCAACUCCUACCCAUUGGUUGGUCAGAUUCA-ACCCUUUUU-GACACGACUAAGAUUCAUAGUU-GCCAGAUAAAAGAUCAGACUUGACUUACCAUGCACUGCG---CUGCUUUU-G----CUCAUGGCCAGUGCAGAAGUUGAAGAGUCACA----------------------- ..((((((........(((((((((((((....-......)))-))).))))))).......))))-))..(((.....))).((((((((((....(((((((.(---((((....-)----)....)))))))))).)))))...)))))...----------------------- ( -44.36, z-score = -3.04, R) >droSim1.chr2R 12761858 144 + 19596830 UAGCAACUCCUACCCAUUGGUUGGUCAGAUUUA-ACCCUUUUU-GACACGACUAAGAUUCAUAGUU-GCCAGAUAAAAGAUCAGACUUGACUUACCAUGCACUGAG---CUGCUUUU-G----CUCAUGGACAGUGCAGAAGUUGAAGAGUCACA----------------------- ..((((((........(((((((((((((....-......)))-))).))))))).......))))-))..(((.....))).((((((((((....(((((((((---(.......-)----)))......)))))).)))))...)))))...----------------------- ( -40.66, z-score = -2.35, R) >droSec1.super_1 11520616 144 + 14215200 UAGCAACUCCUACCCAUUGGUUGGUCACAUUUA-ACCCUUUUU-GACACGACUAAGAUUCAUAGUU-GCCAGAUAAAGGAUCAGACUUGACUUACCAUGCACUGCG---CUGCUUUU-G----CUCAUGGCCAGUGCAGAAGUUGAAGAGUCACA----------------------- ..((((((........(((((((((((......-........)-))).))))))).......))))-))..(((.....))).((((((((((....(((((((.(---((((....-)----)....)))))))))).)))))...)))))...----------------------- ( -41.20, z-score = -2.14, R) >droYak2.chr2R 5983684 145 - 21139217 UAGCAACUUCAACCCAUUGGUUGGUCAGAUUUA-ACCCUUUUUUGACACGACUAAGAUUCAUAGUU-GCCAGAUAAAAGAUCAGACUUGACUUACCAUGCACUACG---CUGCUUUU-G----CUUAUGGCCAGUGCAGAAGUUAAAGAGUCACA----------------------- ..((((((........(((((((((((((....-.......)))))).))))))).......))))-))..(((.....))).((((((((((....((((((..(---((((....-)----)....))).)))))).)))))...)))))...----------------------- ( -40.36, z-score = -3.04, R) >droEre2.scaffold_4845 8250402 144 + 22589142 UAGCAACUUCUACCCAUUGGUUGGUCAGAUUUA-ACUCUUUUU-GACACGACUAUGAUUCAUAGUU-GCCAGAUUAAACAUCAGUCUUGACUUACCAUGCACUACG---CUGCUUUU-A----UUUAUGGCGAGUGCAGAAGUUGAAGAGUCACA----------------------- ...((((((((......((((.(((((((((..-......(((-(...((((((((...)))))))-).)))).........)))).))))).)))).(((((.((---((......-.----.....)))))))))))))))))..........----------------------- ( -42.43, z-score = -3.56, R) >droAna3.scaffold_13266 2774450 141 + 19884421 UGGCAACUUCUACCCAUUGGUUGGUCGAAUUAAU-CCCUUUUUCGACACGACUAAGAUUCAAAGUU-GCAAAAUAAAAGGCCAGACUUGACUUACCAUGUACUAGA---CUGCCCUCCA----GGU-----CAGUGCAGAAGUAGAAGAGUCACA----------------------- ..(((((((.......(((((((((((((.....-......)))))).)))))))......)))))-))..............(((((.((((....((((((.((---((........----)))-----))))))).))))....)))))...----------------------- ( -43.12, z-score = -3.34, R) >dp4.chr3 6631736 147 + 19779522 UAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCGUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAUCAAAGUCCAGGCUUUACUUACCAUGCACUAGG---CUGCUCUUCG----AGCAUAGCCAGGGCAGAAGUAGAAGAGUCACA----------------------- ..(((((((.......((((((((((((....((....))..)))).))))))))......)))))-))..............((((((((((....(((.((.((---((((((...)----)))..)))).))))).)))))...)))))...----------------------- ( -42.92, z-score = -1.84, R) >droPer1.super_2 6851855 147 + 9036312 UAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCAUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAUCAAAGUCCAGGCUUUACUUACCAUGCACUAGG---CUGCUCUUCG----AGCAUAGCCAGGGCAGAAGUAGAAGAGUCACA----------------------- ..(((((((.......((((((((((((..............)))).))))))))......)))))-))..............((((((((((....(((.((.((---((((((...)----)))..)))).))))).)))))...)))))...----------------------- ( -41.26, z-score = -1.61, R) >droWil1.scaffold_180699 1811346 163 + 2593675 UGGCAACUUCUACCCAUUGGUUGGUCAAAU-UA--AUUUAUUUUGAUACGACUAAGAUUCAAAGUUUGCUAGAUCAAUU----AACGUGACUUACCGUGUGAUUGGGCUUUACUUUG------AUAGCUAUU--CACAGAAGUAGAAGCGUAACAGUUGAAUACAGAAGGAGGGAAUA .(....).....(((..((((((((((((.-..--......)))))).))))))((..(((((((..(((.(((((...----.(((........))).))))).)))...))))))------)...))...--.......(((..(((......)))...))).......))).... ( -36.60, z-score = -0.66, R) >droVir3.scaffold_12875 8663867 134 + 20611582 UGGCAACUUCUACCCAUUGGUUGGUCAAAAGUC--UUAUAUUUUGAGCUGACUAAGAUUCAAAGUU-GCAAGAAUAAU------ACGUGACUUACCGUGUAGAUGAG----GCACCG------AUUCUUAUU--UACAGAAGUAGAAGCGUUACA----------------------- ((((..(((((((.....(((.(((((....((--((..((((((((((.....)).)))))))).-..)))).....------...))))).))).((((((((((----(.....------..)))))))--))))...))))))).))))..----------------------- ( -36.70, z-score = -2.56, R) >droMoj3.scaffold_6496 8328870 133 - 26866924 UGGCAACUUAUACCCAUUGGUUGGUCAAAU-UA--UUAUUUUUUGAGACAAUUAAGAUUUAAAGUU-GCCAAAUCAAAU----AACGAAGUUUACCAUGUAAAUGU------CUCCA------AUUAUCAUU--UACAAAAACAGAAGCGUUACA----------------------- (((((((((.......(((((((.(((((.-..--......)))))..)))))))......)))))-)))).......(----((((..........((((((((.------.....------.....))))--))))..........)))))..----------------------- ( -29.67, z-score = -3.29, R) >droGri2.scaffold_15245 2065530 147 + 18325388 UGGCAACUUCUACCCAUUGGUUGGUCAAAAAAUAAUCCAUUUU-GAGCCGACUAAGAUUCAAAGUU-GCUAGAGAAA------UACUUAACUUACCAUGCGUUAAGGCUCUGCCCUUGGGCAUUGAAUAGCUAGCGCAGAAGUAGAAGAGAUACA----------------------- (((((((((.......((((((((((((((.........))))-)).))))))))......)))))-))))......------..(((.((((....((((((..((((.((((....))))......)))))))))).))))..))).......----------------------- ( -42.12, z-score = -2.39, R) >consensus UAGCAACUUCUACCCAUUGGUUGGUCAAAUUUA_ACCCUUUUUUGACACGACUAAGAUUCAAAGUU_GCCAGAUAAAAGAUCAGACUUGACUUACCAUGCACUAGG___CUGCUCUU_G____AUCAUGGCCAGUGCAGAAGUAGAAGAGUCACA_______________________ ..((((((........((((((((((..................))).))))))).......)))).))..............(((((.((((....((((.................................)))).))))....))))).......................... (-16.57 = -15.68 + -0.89)
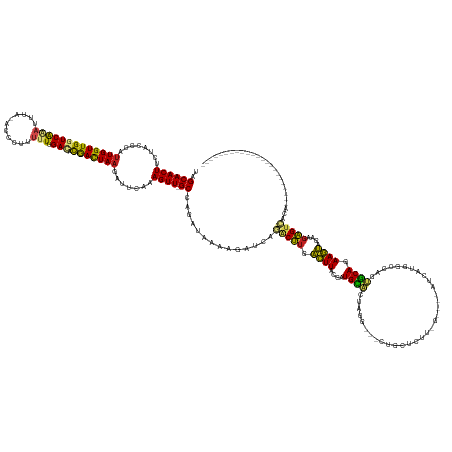
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:29 2011