| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,030,503 – 14,030,618 |
| Length | 115 |
| Max. P | 0.750185 |

| Location | 14,030,503 – 14,030,618 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 71.38 |
| Shannon entropy | 0.61530 |
| G+C content | 0.40461 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -9.75 |
| Energy contribution | -10.03 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
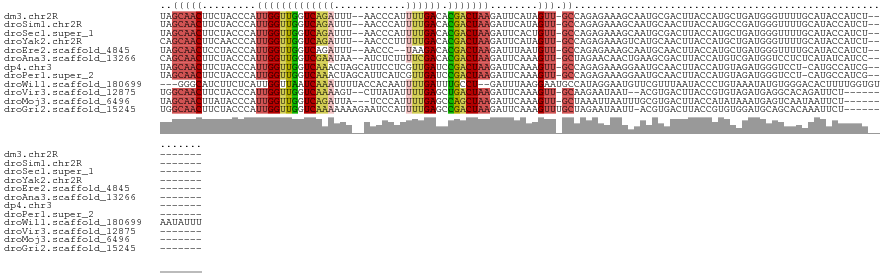
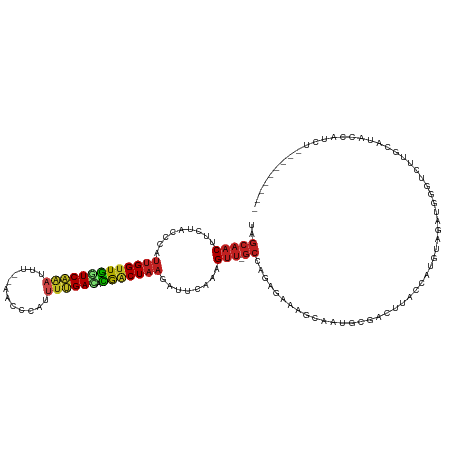
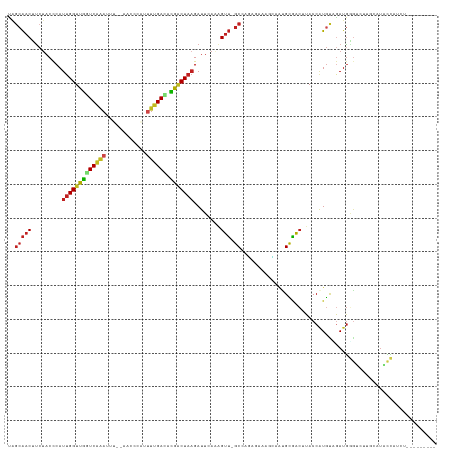
>dm3.chr2R 14030503 115 + 21146708 UAGCAACUUCUACCCAUUGGUUGGUCAGAUUU--AACCCAUUUUGACACGACUAAGAUUCAUAGUU-GCCAGAGAAAGCAAUGCGACUUACCAUGCUGAUGGGUUUUGCAUACCAUCU--------- ..((((((........(((((((((((((...--.......)))))).))))))).......))))-))...........((((((....((((....))))...)))))).......--------- ( -29.36, z-score = -2.10, R) >droSim1.chr2R 12761472 115 + 19596830 UAGCAACUUCUACCCAUUGGUUGGUCAGAUUU--AACCCAUUUUGACACGACUAAGAUUCAUAGUU-GCCAGAGAAAGCAAUGCAACUUACCAUGCCGAUGGGUUUUGCAUACCAUCU--------- ..((((.....((((((((((((((.((....--......(((((...((((((.......)))))-).)))))...((...))..)).)))).)))))))))).)))).........--------- ( -30.70, z-score = -2.82, R) >droSec1.super_1 11520423 115 + 14215200 UAGCAACUUCUACCCAUUGGUUGGUCAGAUUU--AACCCAUUUUGACACGACUAAGAUUCACUGUU-GCCAGAGAAAGCAAUGCGACUUACCAUGCUGAUGGGUUUUGCAUACCAUCU--------- ..((((.....((((((((((((((((((...--.......)))))).)))))....(((.(((..-..))).)))((((.((........))))))))))))).)))).........--------- ( -28.10, z-score = -1.62, R) >droYak2.chr2R 5983090 115 - 21139217 CAGCAACUUCAACCCAUUGGUUGGUCAGAUUU--AACCCUUUUUGACACGACUAAGAUUCAUAGUU-GCCAGAGAAAGUCAUGCAACUUACCAUGCUGAUGGGUUUUGCAUACCAUCU--------- ..((((((........(((((((((((((...--.......)))))).))))))).......))))-))...(((..((.((((((....((((....))))...))))))))..)))--------- ( -31.36, z-score = -2.53, R) >droEre2.scaffold_4845 8250206 113 + 22589142 UAGCAACUCCUACCCAUUGGUUGGUCAGAUUU--AACCC--UAAGACACGACUAAGAUUUAAUGUU-GCCAGAGAAAGCAAUGCAACUUACCAUGCUGAUGGGUUUUGCAUACCAUCU--------- ..((((.....((((((..((((((.((..((--((..(--(.((......)).))..)))).(((-((........)))))....)).)))).))..)))))).)))).........--------- ( -27.10, z-score = -1.52, R) >droAna3.scaffold_13266 2774294 115 + 19884421 CAGCAACUUCUACCCAUUGGUUGGUCGAAUAA--AUCUCUUUUCGACACGACUAAGAUUCAAAGUU-GCUAGAACAACUGAAGCGACUUACCAUGUCGAUGGUCCUCUCAUAUCAUCC--------- .((((((((.......(((((((((((((...--.......)))))).)))))))......)))))-)))..............(((.......)))((((((........)))))).--------- ( -30.62, z-score = -3.18, R) >dp4.chr3 6631568 116 + 19779522 UAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCCUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAGAAAGGAAUGCAACUUACCAUGUAGAUGGGUCCU-CAUGCCAUCG--------- ..(((......(((((((...((((.......((((((((..(((...(((((.........))))-).)))....)))))))).....))))....)))))))...-..))).....--------- ( -29.30, z-score = -1.25, R) >droPer1.super_2 6851687 116 + 9036312 UAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCAUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAGAAAGGAAUGCAACUUACCAUGUAGAUGGGUCCU-CAUGCCAUCG--------- ..(((......(((((((...((((.......((((((.((.(((...(((((.........))))-).))).))...)))))).....))))....)))))))...-..))).....--------- ( -26.20, z-score = -0.37, R) >droWil1.scaffold_180699 1811180 122 + 2593675 ---GGGCAUCUUCUCAUUGGUUAAUCAAAUUUUACCACAAUUUUGAUUUGCCU--GAUUUAAGGAAUGCCAUAGGAAUGUUCGUUUAAUACCCUGUAAAUAUGUGGGACACUUUUGGUGUAAUAUUU ---.((((((((......(((.(((((((............))))))).))).--......))).)))))....(((((((.........(((...........)))((((.....))))))))))) ( -25.02, z-score = -0.17, R) >droVir3.scaffold_12875 8663702 109 + 20611582 UGGCAACUUCUACCCAUUGGUUGGUCAAAAGU--CUUAUAUUUUGAGCUGACUAAGAUUCAAAGUU-GCAAGAAUAAU--ACGUGACUUACCGUGUAGAUGAGGCACAGAUUCU------------- ..(((((((.......(((((..(((((((..--......)))))).)..)))))......)))))-)).(((((...--..(((.((((.(.....).)))).)))..)))))------------- ( -27.32, z-score = -1.49, R) >droMoj3.scaffold_6496 8328701 110 - 26866924 UAGCAACUUAUACCCAUUGGUUGGUCAGAUUA---UCCCAUUUUGAGCCAGCUAAGAUUCAAAGUU-GCUAAAUUAAUUUGCGUGACUUACCAUAUAAAUGAGUCAAUAAUUCU------------- (((((((((.......(((((((((((((...---......))))).))))))))......)))))-))))............(((((((.........)))))))........------------- ( -29.02, z-score = -3.97, R) >droGri2.scaffold_15245 2065361 113 + 18325388 UGGCAACUUCUACCCAUUGGUUGGUCAAAAAAAGAAUCCAUUUUGAGCCGACUAAGAUUCAAAGUUUGCUAGAAUAAUU-ACGUGACUUACCGUGUGGAUGCAGCACAAAUUCU------------- (((((((((.......((((((((((((((..........)))))).))))))))......))).))))))........-............((((.......)))).......------------- ( -22.92, z-score = -0.24, R) >consensus UAGCAACUUCUACCCAUUGGUUGGUCAAAUUU__AACCCAUUUUGACACGACUAAGAUUCAAAGUU_GCCAGAGAAAGCAAUGCGACUUACCAUGUAGAUGGGUCUUGCAUACCAUCU_________ ..(((((.........(((((((((((((............)))))).)))))))........))).)).......................................................... ( -9.75 = -10.03 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:28 2011