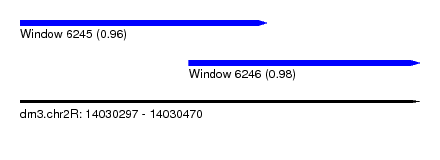
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,030,297 – 14,030,470 |
| Length | 173 |
| Max. P | 0.981602 |

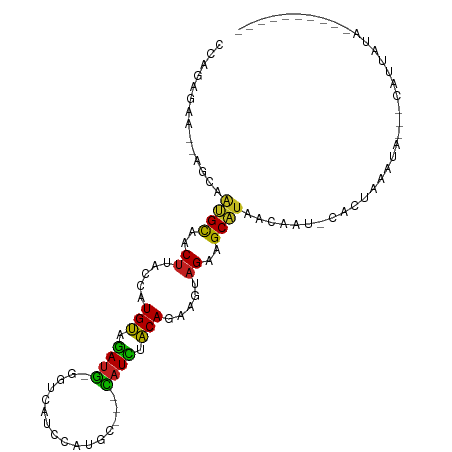
| Location | 14,030,297 – 14,030,404 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.54645 |
| G+C content | 0.39441 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.08 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14030297 107 + 21146708 UUCAUAUCUCGCAACUCCUACCCAUUGGUUGGUCAGAUU--UAUCUCUUUUUGACACGACUAAGAUUCAAUGUU-GCCAGAGAAAGCAA--UGCAACUUACCAUGUAGAUGG--- ......(((((((((.........(((((((((((((..--........)))))).)))))))........)))-))..))))......--.........((((....))))--- ( -24.53, z-score = -1.61, R) >droSim1.chr2R 12761273 107 + 19596830 UACAAAUCUCGCAACUCCUACCCAUUGGUUGGUCAGAUU--UAUCUCUUUUUGACACGACUAAGAUUCAAUGUU-GCCAGAGAAAGCGA--UGCAACUUACCAUGUAGAUGG--- ......(((((((((.........(((((((((((((..--........)))))).)))))))........)))-))..))))......--.........((((....))))--- ( -24.53, z-score = -1.54, R) >droSec1.super_1 11520220 107 + 14215200 UACAAAUCUCGCAACUCCUACCCAUUGGUUGGUCAGAUU--UAUCUCUUUUUGACACGACUAAGAUUCAAUGUU-GCCAGAGAAACCGA--UGCAACUUACCAUGUAGAUGG--- ......(((((((((.........(((((((((((((..--........)))))).)))))))........)))-))..))))..(((.--((((........))))..)))--- ( -25.43, z-score = -2.24, R) >droYak2.chr2R 5982887 104 - 21139217 ----UAUGUCGCAACUCCUACCCAUUGGUUGGUCAGAUU--UAACCCUUUUUGACACGACUAAGAUUAACUGUUUGCCAGAGAAAGUCA--UGCAACUUACCAUGUAGAUGG--- ----...((.(((...........(((((((((((((..--........)))))).)))))))((((..(((.....)))....)))).--))).))...((((....))))--- ( -20.30, z-score = -0.30, R) >droEre2.scaffold_4845 8249026 107 + 22589142 UAUAUAUCUCGCAACUUCUACCCAUUGGUUGGUCAGAUU--UAACGCUUUUUGACACGACUAAGAUUUAAUGUU-GCCAGAGAAAGCAA--UGCAACUUACCAUGCUGAUGG--- ......(((((((((.........(((((((((((((..--........)))))).)))))))........)))-))..)))).((((.--((........)))))).....--- ( -25.03, z-score = -2.10, R) >droAna3.scaffold_13266 2774146 99 + 19884421 --------AGGCAGCCUCUACCCAUUGGUUGGUCAAUUUU-UCCACAUUUUUGACACGACUAAGAUUCAAGGCU-GCCAGAAUAACUAG---GCGACUUACCAUGCCAAUGG--- --------.((((((((.......((((((((((((....-.........))))).)))))))......)))))-)))..........(---(((........)))).....--- ( -32.54, z-score = -4.04, R) >dp4.chr3 6631392 109 + 19779522 GUCCUCUAUAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCCUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAGAAAGGAA--UGCAACUUACCAUGUAGAUGG--- .((((((...(((((((.......((((((((((((...((.....))..)))).))))))))......)))))-))...)))..))).--.........((((....))))--- ( -26.52, z-score = -1.39, R) >droPer1.super_2 6851679 109 + 9036312 AUAAUAAAUAGCAACUUCUACCCAUUGGUUGGUCAAACUAGCAUUCAUCGUUGAUCCGACUAAGAUUCAAAGUU-GCCAGAGAAAGGAA--UGCAACUUACCAUGUAGAUGG--- ..............(.(((((...((((((((((((..............)))).))))))))......(((((-((............--.))))))).....))))).).--- ( -21.06, z-score = -0.39, R) >droWil1.scaffold_180699 1811006 110 + 2593675 UACAAACUUCGCAACUUCUACCCAUUGGUUGGUCAAAU---UAAUUUAUUUUGAUACAACUAAGAUUCAAAGUUUGCUAGAUCAAUGUA--CGUGACUUACCAUGUGAAUGGAAU ..........(((((((.......(((((((((((((.---........)))))).)))))))......))).))))....((....((--((((......))))))....)).. ( -21.42, z-score = -1.22, R) >droVir3.scaffold_12875 8663534 101 + 20611582 -----UACAAGCAACUUCUACCCAUUGGUUGGUCAAAAG--UCUUAUAUUUUGAGCUGACUAAGAUUCAAAGUU-GCAAGAAUAAUA----CGUGACUUACCGUGUAGAUGAG-- -----.....(((((((.......(((((..(((((((.--.......)))))).)..)))))......)))))-)).......(((----((........))))).......-- ( -23.62, z-score = -1.82, R) >droMoj3.scaffold_6496 8328530 102 - 26866924 -----GCUAGGCAACUUAUACCCAUUGGUUGGUCAGAU---UAUCCCAUUUUGAGCCAGCUAAGAUUCAAAGUU-GCUAAAUUAAUUUG--CGUGACUUACCAUGUAAAUAAG-- -----....((((((((.......(((((((((((((.---........))))).))))))))......)))))-)))......(((((--((((......)))))))))...-- ( -30.52, z-score = -4.07, R) >droGri2.scaffold_15245 2065178 104 + 18325388 -----GACAAGCAACUUCUACCCAUUGGUUGGUCAAAAA--UCCUUCAUUUUGAGUCAACUAAGAUUCAUUGUU-GCUAGAUUAUUAUUUCCGUGACUUACCAUGUAGAUGG--- -----....((((((.........((((((((((((((.--.......)))))).))))))))........)))-))).......(((((.((((......)))).))))).--- ( -23.33, z-score = -1.87, R) >consensus ___AUAUCUCGCAACUUCUACCCAUUGGUUGGUCAAAUU__UAUCCCUUUUUGACACGACUAAGAUUCAAUGUU_GCCAGAGAAAGCAA__UGCAACUUACCAUGUAGAUGG___ ..........(((((.........((((((((((((..............))))).)))))))........))).)).......................((((....))))... (-12.52 = -12.08 + -0.44)

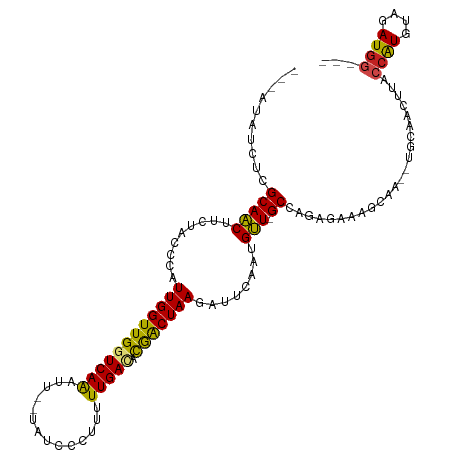
| Location | 14,030,370 – 14,030,470 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.78533 |
| G+C content | 0.37433 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -7.64 |
| Energy contribution | -6.22 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14030370 100 + 21146708 CCAGAGAA--AGCAAUGCAACUUACCAUGUAGAUG-GCUGAUCUAUGC---CAUCCACAGAAGUAGAAGCAUAACAAA-CACUAAAUAAUACAUCAUAUG-AUUAGAC ........--....((((..((.....(((.((((-((........))---)))).))).....))..))))......-..((((...(((.....))).-.)))).. ( -18.70, z-score = -2.05, R) >droSim1.chr2R 12761346 97 + 19596830 CCAGAGAA--AGCGAUGCAACUUACCAUGUAGAUG-GUCGAUCUAUGC---CAUCCACAGAAGUAGAAGCAUAACAAA-CACUAAAUG---CAUCAUAUG-AUUAGAC ........--...((((((((((....(((.((((-((........))---)))).))).))))....(.....)...-.......))---)))).....-....... ( -18.70, z-score = -1.61, R) >droSec1.super_1 11520293 97 + 14215200 CCAGAGAA--ACCGAUGCAACUUACCAUGUAGAUG-GUCGAUCUAUGC---CAUCCACAGAAGUAGAAGCAUUACAAA-CACUAAAUG---CAUCAUAUG-AUUAGAC ........--...((((((........(((.((((-((........))---)))).)))...((((.....))))...-.......))---)))).....-....... ( -18.90, z-score = -1.90, R) >droYak2.chr2R 5982957 98 - 21139217 CCAGAGAA--AGUCAUGCAACUUACCAUGUAGAUG-GUCGAUCCAUAU---CAUUUACAGAAGUAGAAGCAUAACAAA-CACUAAAUG---CAUCAUCGAUAUUAGAC ........--.((.((((..((.....((((((((-((........))---)))))))).....))..)))).))...-..((((...---(......)...)))).. ( -16.90, z-score = -1.45, R) >droEre2.scaffold_4845 8249099 97 + 22589142 CCAGAGAA--AGCAAUGCAACUUACCAUGCUGAUG-GGUUUUGCAAAC---CAUCUGCAGAAGUAGAAGCAUAACAAA-CACUAAAUG---CAUUAUACG-AUUAGGU ........--.....(((((....((((....)))-)...))))).((---(.(((((....))))).((((......-......)))---)........-....))) ( -17.80, z-score = -0.11, R) >droAna3.scaffold_13266 2774212 82 + 19884421 CCAGAAUA--ACUAG-GCGACUUACCAUGCCAAUG-GUUUCCCUGUAC---CAUUCGCAGAAGUUGAAGCCUAACACU-UAGGAAACUUG------------------ ........--..(((-(((((((....(((.((((-((........))---)))).))).)))))...))))).....-.((....))..------------------ ( -21.70, z-score = -2.82, R) >dp4.chr3 6631467 84 + 19779522 CCAGAGAA--AGGAAUGCAACUUACCAUGUAGAUG-GGUCCU-CAUGC---CAUCGACAGAAGUAGAAGCAUUACAUU-CA--GAAUA---UCUUGU----------- .....(((--.(.(((((..((.....(((.((((-(.....-....)---)))).))).....))..))))).).))-).--.....---......----------- ( -17.10, z-score = -0.59, R) >droPer1.super_2 6851754 84 + 9036312 CCAGAGAA--AGGAAUGCAACUUACCAUGUAGAUG-GGUCCU-CAUGC---CAUCGACAGAAGUAGAAGCAUUACAUU-CA--GAAUA---UCUUGA----------- .....(((--.(.(((((..((.....(((.((((-(.....-....)---)))).))).....))..))))).).))-).--.....---......----------- ( -17.10, z-score = -0.59, R) >droWil1.scaffold_180699 1811079 83 + 2593675 CUAGAUCA--AUGUACGUGACUUACCAUGUGAAUGGAAUUAUUCACUCAACUAUUUACAGAAGAAGAAGCGUAACAAUUUACUCU----------------------- ........--.(((((((..(((....(((((((((..............)))))))))....)))..)))).))).........----------------------- ( -15.84, z-score = -1.56, R) >droVir3.scaffold_12875 8663602 89 + 20611582 CAAGAAUA--AU--ACGUGACUUACCGUGUAGAUG-AGGCACAGAUUC--UUAUUUACAGAAGUAGAAGCGUUACAAU-U--UAGUAAAUUUUUGAGAA--------- (((((((.--..--.(((.((((....((((((((-(((.......))--))))))))).))))....)))((((...-.--..)))))))))))....--------- ( -18.80, z-score = -2.48, R) >droMoj3.scaffold_6496 8328597 93 - 26866924 CUAAAUUA--AUUUGCGUGACUUACCAUGUAAAUA-AGUCAUUAACUC--UUAUUUACAGAAGUAGAAGCGUUACACUAU--UAAUUAAGAUAUUAGAAU-------- ((.(((((--((..((((.((((....((((((((-((.........)--))))))))).))))....))))......))--))))).))..........-------- ( -18.90, z-score = -2.81, R) >droGri2.scaffold_15245 2065246 102 + 18325388 CUAGAUUAUUAUUUCCGUGACUUACCAUGUAGAUG-GG-CAGCAAUUC--CUUUUUACAGAAGUAGAAGCGUUACAAUCUACUACAAUAGAUGCUAUGGUUAAGAA-- ....................(((((((((((((.(-((-.(....).)--)).))))).(.((((((..........)))))).)..........)))).))))..-- ( -18.00, z-score = 0.12, R) >consensus CCAGAGAA__AGCAAUGCAACUUACCAUGUAGAUG_GGUCAUCCAUGC___CAUCUACAGAAGUAGAAGCAUAACAAU_CACUAAAUA___CAUUAUA__________ ..............((((..((.....(((.((((................)))).))).....))..)))).................................... ( -7.64 = -6.22 + -1.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:27 2011