
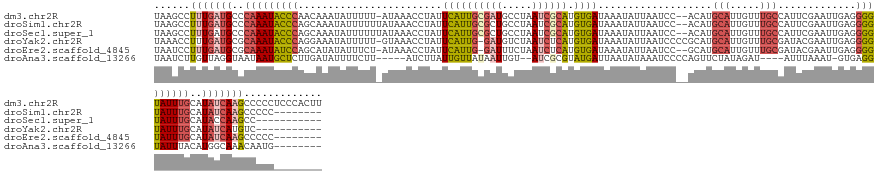
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,977,197 – 13,977,342 |
| Length | 145 |
| Max. P | 0.731326 |

| Location | 13,977,197 – 13,977,342 |
|---|---|
| Length | 145 |
| Sequences | 6 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Shannon entropy | 0.42542 |
| G+C content | 0.35254 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -17.66 |
| Energy contribution | -19.47 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
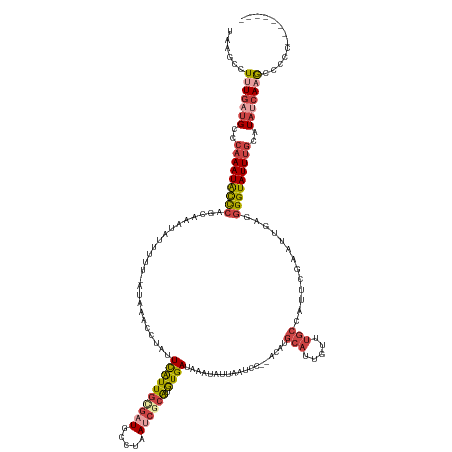
>dm3.chr2R 13977197 145 + 21146708 UAAGCCUUUGAUGCCCAAAUACCCAACAAAUAUUUUU-AUAAACCUAUUCAUUGCGAUGCCUAAUCGCAUGUGAUAAAUAUUAAUCC--ACAUGCAUUGUUUGCCAUUCGAAUUGAGGGGUAUUUGCAUAUCAAGCCCCCUCCCACUU ...((..((((((..(((((((((.....(((....)-)).......((((...(((.((.((((.(((((((.............)--))))))))))...))...)))...)))))))))))))..))))))))............ ( -33.52, z-score = -3.03, R) >droSim1.chr2R 12710332 138 + 19596830 UAAGCCUUUGAUGCCCAAAUACCCAGCAAAUAUUUUUUAUAAACCUAUUCAUUGCGCUGCCUAAUCGCAUGUGAUAAAUAUUAAUCC--ACAUGCAUUGUUUGCCAUUCGAAUUGAGGGGUAUUUGCAUAUCAAGCCCCC-------- ...((..((((((..(((((((((..((((((.....))).......(((..((.((....((((.(((((((.............)--))))))))))...))))...))))))..)))))))))..))))))))....-------- ( -32.92, z-score = -2.28, R) >droSec1.super_1 11470882 135 + 14215200 UAAGCCUUUGAUGCCCAAAUACCCAGCAAAUAUUUUUUAUAAACCUAUUCAUUGCGCUGCCUAAUCGCAUGUGAUAAAUAUUAAUCC--ACAUGCAUUGUUUGCCAUUCGAAUUGAGGGGUAUUUGCAUACCAAGCC----------- ...((.....((((..((((((((..((((((.....))).......(((..((.((....((((.(((((((.............)--))))))))))...))))...))))))..)))))))))))).....)).----------- ( -29.42, z-score = -1.26, R) >droYak2.chr2R 5933166 135 - 21139217 UAAACCUUUGAUGCGCAAAUACCCAGGAAAUAUUUUU-GUAAACCUAUUCAUUG-GAUGUCUAAUCUCAUGUGAUAAAUAUUAAUCCCCGCAUGCAUUGUUUGCGAUACGAAUUGAGGGGUAUUUGCAUAUCAUGUC----------- ........(((((.((((((((((...........((-((((((......((((-(....)))))..((((((...............))))))....))))))))...........)))))))))).)))))....----------- ( -34.21, z-score = -2.09, R) >droEre2.scaffold_4845 8199459 136 + 22589142 UAAUCCUUUGAUGCGCAAAUAUCCAGCAUAUAUUUCU-AUAAACCUAUUCAUUG-GAUUUCUAAUCUCAUGUGAUAAAUAUUAAUCC--GCAUGCAUUGUUUGCGAUACGAAUUGAGGGGUAUUUGCAUAUCAAGCCCCC-------- ......(((((((.((((((((((..((.........-.(((((..((.(((((-((((..(((((......)))...))..)))))--).))).)).)))))..........))..)))))))))).))))))).....-------- ( -30.15, z-score = -1.28, R) >droAna3.scaffold_13266 9665136 128 - 19884421 UAAUCUUGUUAGGUAAUAAUGCUCUUGAUAUUUUCUU-----AUCUUAUUGUUAUAAUUGU--AUCGCGUAUGAUUAAUAUAAAUCCCCAGUUCUAUAGAU----AUUUAAAU-GUGAGGUAUUUACAUGGCAAACAAUG-------- .....((((((.((((..((((.(((.((((((...(-----((((.(((((((((.(((.--((((....)))))))))))).....)))).....))))----)...))))-))))))))))))).))))))......-------- ( -18.10, z-score = 0.72, R) >consensus UAAGCCUUUGAUGCCCAAAUACCCAGCAAAUAUUUUU_AUAAACCUAUUCAUUGCGAUGCCUAAUCGCAUGUGAUAAAUAUUAAUCC__ACAUGCAUUGUUUGCCAUUCGAAUUGAGGGGUAUUUGCAUAUCAAGCCCCC________ ......(((((((..(((((((((........................((((((((((.....)))))).))))...................(((.....))).............)))))))))..)))))))............. (-17.66 = -19.47 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:24 2011