
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,971,868 – 13,971,963 |
| Length | 95 |
| Max. P | 0.848090 |

| Location | 13,971,868 – 13,971,963 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 55.99 |
| Shannon entropy | 0.67848 |
| G+C content | 0.43577 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -5.43 |
| Energy contribution | -9.05 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13971868 95 + 21146708 --AAAUUAAGAUGCGCGGCUUAUGUACG--GUUGGUCACUGACACCGUGUAUAUAAGAAGACAAAUACCUGCUCUGCAACGGCAGAAAAGGGAAAUGAA --........((((((((....(((.((--((.....)))))))))))))))...............(((..(((((....)))))..)))........ ( -25.20, z-score = -1.96, R) >droYak2.chrU 7648380 96 + 28119190 AAAAAAAACAAUGGGCGAGGUAUAUAUG--GUCGGCCACUGACACCGUGUAUAUAACA-GACAAAUAACUGUUCUGCAACUGCACAAAAGGGAAACGAA ...........((.((.(((((((((((--((.(........))))))))))))..((-((((......)).))))...)))).))....(....)... ( -19.40, z-score = -0.96, R) >dp4.Unknown_group_140 12030 95 + 24112 --AACACAAAUUAUGCUGCCAAUGCACACAACCAAACGCCGCCAAAUUAUGCACAAUUUGGCG--CAACGAUAGCGUUAUCGAAGUAAAGGCAGCACCA --...........(((((((..(((............((.((((((((......)))))))))--)..(((((....)))))..)))..)))))))... ( -30.00, z-score = -4.09, R) >droPer1.super_158 13903 95 - 72041 --AACACAAAUUAUGCUGCCAAUGCACACAACCAAAUGCCGCCAAAUUAUGCACAAUUUGGCG--CAACGAUAGCGUUACCGAAGUAAAGGCAGCACCA --...........(((((((..(((............((.((((((((......)))))))))--)((((....))))......)))..)))))))... ( -28.40, z-score = -3.31, R) >consensus __AAAACAAAAUAUGCGGCCAAUGCACA__ACCAAACACCGACAAAGUAUACACAAUAUGACA__CAACGAUACCGCAACCGAAGAAAAGGCAAAACAA .............(((((((...................(((((((((......))))))))).....(((((....))))).......)))))))... ( -5.43 = -9.05 + 3.62)

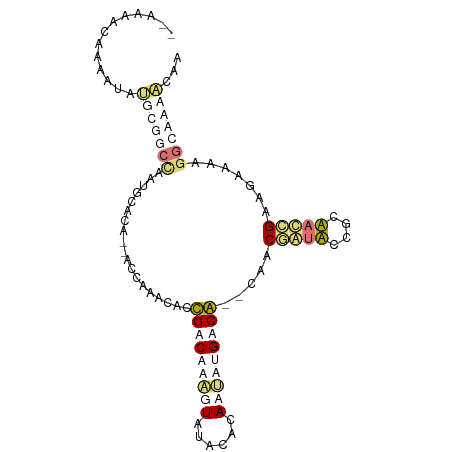

| Location | 13,971,868 – 13,971,963 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 55.99 |
| Shannon entropy | 0.67848 |
| G+C content | 0.43577 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -8.20 |
| Energy contribution | -9.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13971868 95 - 21146708 UUCAUUUCCCUUUUCUGCCGUUGCAGAGCAGGUAUUUGUCUUCUUAUAUACACGGUGUCAGUGACCAAC--CGUACAUAAGCCGCGCAUCUUAAUUU-- ........(((.((((((....)))))).)))....(((...(((((....((((((((...)))..))--)))..)))))....))).........-- ( -18.60, z-score = -1.03, R) >droYak2.chrU 7648380 96 - 28119190 UUCGUUUCCCUUUUGUGCAGUUGCAGAACAGUUAUUUGUC-UGUUAUAUACACGGUGUCAGUGGCCGAC--CAUAUAUACCUCGCCCAUUGUUUUUUUU ............(((((.....(((((.(((....)))))-)))......)))))...((((((.(((.--..........))).))))))........ ( -17.30, z-score = -0.75, R) >dp4.Unknown_group_140 12030 95 - 24112 UGGUGCUGCCUUUACUUCGAUAACGCUAUCGUUG--CGCCAAAUUGUGCAUAAUUUGGCGGCGUUUGGUUGUGUGCAUUGGCAGCAUAAUUUGUGUU-- ..((((((((..(((..(((((((((........--((((((((((....)))))))))))))))..)))).)))....))))))))..........-- ( -32.60, z-score = -1.73, R) >droPer1.super_158 13903 95 + 72041 UGGUGCUGCCUUUACUUCGGUAACGCUAUCGUUG--CGCCAAAUUGUGCAUAAUUUGGCGGCAUUUGGUUGUGUGCAUUGGCAGCAUAAUUUGUGUU-- ..((((((((..(((....)))((((.((((.((--((((((((((....))))))))).)))..)))).)))).....))))))))..........-- ( -37.70, z-score = -3.34, R) >consensus UGCUGCUCCCUUUACUGCGGUAACACAACAGUUA__CGCCAAAUUAUACACAAGGUGGCAGCGUCCGAC__CGUACAUAGGCAGCACAAUUUGUGUU__ ...(((((((.......(((((....))))).....(((((((((......)))))))))((((........))))...)))))))............. ( -8.20 = -9.70 + 1.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:23 2011