
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,966,671 – 13,966,844 |
| Length | 173 |
| Max. P | 0.546007 |

| Location | 13,966,671 – 13,966,844 |
|---|---|
| Length | 173 |
| Sequences | 6 |
| Columns | 175 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Shannon entropy | 0.46737 |
| G+C content | 0.45250 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -18.26 |
| Energy contribution | -21.08 |
| Covariance contribution | 2.82 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13966671 173 + 21146708 UCUGAAAAACACCUUUGCACAUCCUAUUUGCCUCGCCGGAGCGGAGAAAAUAAAACGUUCAAGUGUCAUUUAUCUCACUGGGCCAAAA-CUUUCCAA-CUGACUUUCCACCGCCCGCACACACACGAAAAAGUGUUCUAACCAAGUGUGAAUGUGCGGGUGAACUCUGCAUAUUU .....................(((.............)))((((((..........(((((.(((..........)))))))).....-........-............(((((((((((((((......((......))...)))))..))))))))))..))))))...... ( -41.32, z-score = -0.93, R) >droSim1.chr2R 12705154 173 + 19596830 GCUGAAAAACACCUCCGCACAUCCUAUUUGCCUCGCCGGAGCGGAGAAAAUAAAACGUUCAAGUGCCAUUUAUCUCACUGGGCCAAAAACUUUCCAAACUGACUUUCCAAAGCCCGCACACAC--GAAAAAGUGUGCUAACCAAGUGUGAAUGUGCGGGUGAACUCUGCAUAUUU ((.((.......((((((...(((.............)))))))))..........(((((..(((((((((...((((((((.........((......)).........))))((((((..--......))))))......)))))))))).)))..))))))).))...... ( -47.19, z-score = -2.21, R) >droSec1.super_1 11465662 173 + 14215200 GCUGAAAAACACCUCCGCACAUCCUAUUUGCCUCGCCGGAGCGGAGAAAAUAAAACGCUCAAGUGCCAUUUAUCUCACUGGGCCAAAAACUUUCCAAACUGACUUUCCACCGCCCGCACACAC--GAAAAAGUGUGCUUACCAAGUGUGAAUGUGCGGGUGAACUCUGCAUAUUU ((.((....((((..(((((((...........(((..(((((............)))))..))).......((.((((((((.........((......)).........))))((((((..--......))))))......)))).)))))))))))))...)).))...... ( -47.97, z-score = -2.36, R) >droYak2.chr2R 5927993 133 - 21139217 ------------------------GAAACACCUUGC---------AUAUAUAAAACGUUCAAGUGCCAUUUAUCUCACUAGGCCAAAA-CUUUCCAA-CUGACUU-CCAU--CCCGCACACGU----AAAAGUGUGCUAACCAAGUGUGAAUGUGCGGGUGAACUCUGCAUAUUU ------------------------....(((((((.---------...........(((.(((((((.............)))....)-)))...))-)......-....--...((((((..----....))))))....)))).)))((((((((((....).))))))))). ( -30.62, z-score = -1.64, R) >droEre2.scaffold_4845 8194479 163 + 22589142 --GCAGAAACACCUACGCAUACAUAAUUUGCCUCGCCAGAGCGGAGAAAAUAAAACGCUCAAGUGCCAUUUAUCUCACUGGGCCAAAA-CUUUCCAA-CUGACUU-CCA---CCCGCACACGA----AAAAGUGUGGUAACCAAGUGAGAAUGUGCGGGUGAACUCUGCAUAUUU --(((((..((((..(((((.............(((..(((((............)))))..))).......(((((((((.......-........-.((....-.))---.((((((....----....))))))...)).))))))))))))..))))...)))))...... ( -49.20, z-score = -3.44, R) >droAna3.scaffold_13266 9660653 152 - 19884421 --ACAAAUACACCUCAACAUGUCGAACUAACCCCACCAGAA--GAGGGAAUAAUAUGUUCAAGUGCCAUUUAUCUCUCUGGGGCCAAAACUUUCCA--UCGACUU-UCACU-GCCGCACACGC---------------AUUUAUGUCUGUAUGCGUGGGUGAACUCUGCAUAUUU --.......((((.......(((((..........(((((.--((((((((..(((......)))..)))).)))))))))((..........)).--)))))..-.....-......(((((---------------((..........))))))))))).............. ( -32.10, z-score = 0.06, R) >consensus __UGAAAAACACCUCCGCACAUCCUAUUUGCCUCGCCGGAGCGGAGAAAAUAAAACGUUCAAGUGCCAUUUAUCUCACUGGGCCAAAA_CUUUCCAA_CUGACUU_CCAC__CCCGCACACAC____AAAAGUGUGCUAACCAAGUGUGAAUGUGCGGGUGAACUCUGCAUAUUU ............((((((......................))))))..........(((((..(((((((((...((((.((..........((......)).............((((((..........))))))...)).)))))))))).)))..)))))........... (-18.26 = -21.08 + 2.82)
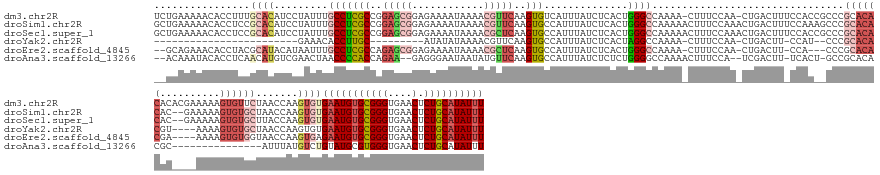
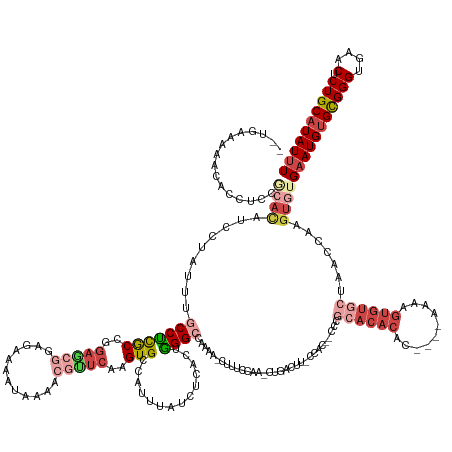

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:21 2011