| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,956,720 – 13,956,852 |
| Length | 132 |
| Max. P | 0.991711 |

| Location | 13,956,720 – 13,956,852 |
|---|---|
| Length | 132 |
| Sequences | 7 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 73.35 |
| Shannon entropy | 0.52427 |
| G+C content | 0.48120 |
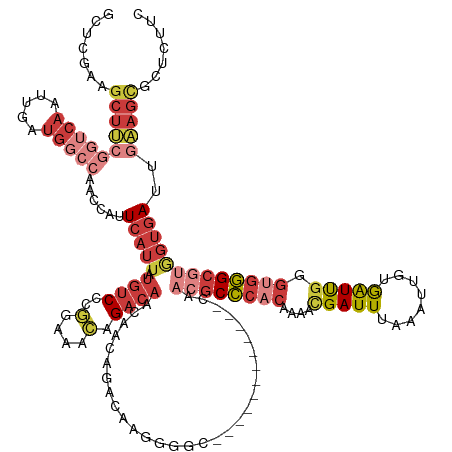
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -21.46 |
| Energy contribution | -23.93 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
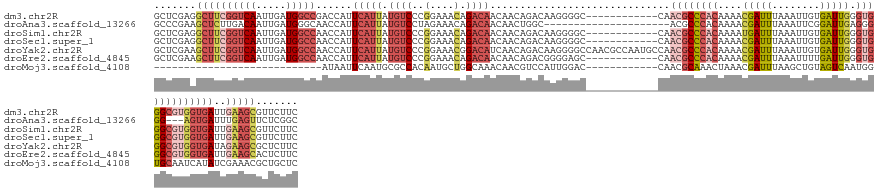
>dm3.chr2R 13956720 132 - 21146708 GCUCGAGGCUUCGGUCAAUUGAUGGCCGACCAUUCAUUAUGUCCCGGAAACAGACAACAACAGACAAGGGGC------------CAACGCCCACAAAACGAUUUAAAUUGUGAUUGGGUGGGCGUGGUGAUUGAAGCGUUCUUC ....(((((((((((((.....(((((((....))....((((..(....).)))).............)))------------))((((((((....(((((........))))).))))))))..)))))))))).)))... ( -48.10, z-score = -2.90, R) >droAna3.scaffold_13266 9650614 120 + 19884421 GCCCGAAGCUCUUGACAAUUGAUGGGCAACCAUUCAUUAUGUCCUAGAAACAGACAACAACUGGC---------------------ACGCCCACAAAACGAUUUAAAUUCGGAUUGAGGGGG---AGUGAUUUGAGUUCUCGGC ..(((((((((..((((..((((((....))).)))...)))).......(((.......))).(---------------------((.(((.(((..(((.......)))..)))...)))---.)))....))))).)))). ( -35.50, z-score = -2.18, R) >droSim1.chr2R 12695264 132 - 19596830 GCUCGAGGCUUCGGUCAAUUGAUGGCCAACCAUUCAUUAUGUCCCGGAAACAGACAACAACAGACAAGGGGC------------CAACGCCCACAAAAUGAUUUAAAUUGUGAUUGGGUGGGCGUGGUGAUUGAAGCGUUCUUC ....(((((((((((((.....(((((............((((..(....).)))).............)))------------))((((((((..(((.((.......)).)))..))))))))..)))))))))).)))... ( -47.31, z-score = -2.88, R) >droSec1.super_1 11455744 132 - 14215200 GCUCGAGGCUUCGGUCAAUUGAUGGCCAACCAUUCAUUAUGUCCCGGAAACAGACAACAACAGACAAGGGGC------------CAACGCCCACAAAACGAUUUAAAUUGUGAUUGGGUGGGCGUGGUGAUUGAAGCGUUCUUC ....(((((((((((((.....(((((............((((..(....).)))).............)))------------))((((((((....(((((........))))).))))))))..)))))))))).)))... ( -47.81, z-score = -3.04, R) >droYak2.chr2R 5918040 144 + 21139217 GCUCGAAGCUUCGGUCAAUUGAUGGCCAACCAUUCAUUAUGUCCCGGAAACGGACAUCAACAGACAAGGGGCCAACGCCAAUGCCAACGCCCACAAAACGAUUUAAAUUGUGAUUGGGUGGGCGUGGUGAUAGAAGCGCUCUUC ....((.(((((((((..(((((((....)))).....((((((.(....))))))).......)))..))))..(((((.((((..((((((....(((((....)))))...)))))))))))))))...)))))..))... ( -53.50, z-score = -3.29, R) >droEre2.scaffold_4845 8184786 132 - 22589142 GCUCGAAGCUUCGGUCAAUUGAUGGCCAACCAUUCAUUAUGUCCCGGAAACAGACAACAACAGACGGGGAGC------------CAACGCCCACAAAACGAUUUAAAUUUUGAUUGGGUGGGCGUGGUGAUUGAAGCACUCUUC .......((((((((((((.(((((....))))).)))...(((((..................))))).((------------((.(((((((....(((((........))))).))))))))))))))))))))....... ( -46.17, z-score = -3.26, R) >droMoj3.scaffold_4108 4607 104 + 6700 ----------------------------AUAAUUCAAUGCGCCACAAUGCUGGCAAACAACGUCCAUUGGAC------------CAACGCAAACUAAACGAUUUAAGCUGUAGUCAAUGGUGCAAUCAUAUCGAAACGCUGCUC ----------------------------.........(((((((...(((((.....))..((((...))))------------....)))........((((........))))..))))))).................... ( -18.30, z-score = 0.76, R) >consensus GCUCGAAGCUUCGGUCAAUUGAUGGCCAACCAUUCAUUAUGUCCCGGAAACAGACAACAACAGACAAGGGGC____________CAACGCCCACAAAACGAUUUAAAUUGUGAUUGGGUGGGCGUGGUGAUUGAAGCGCUCUUC .......((((((((((.....))))).(((........((((..(....).))))..............................((((((((....(((((........))))).)))))))))))....)))))....... (-21.46 = -23.93 + 2.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:20 2011