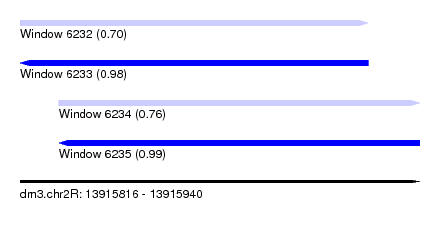
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,915,816 – 13,915,940 |
| Length | 124 |
| Max. P | 0.991852 |

| Location | 13,915,816 – 13,915,924 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Shannon entropy | 0.11687 |
| G+C content | 0.62795 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -36.60 |
| Energy contribution | -36.27 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13915816 108 + 21146708 GCGCCAAGGAGCUGGCAGGCAGGAUGGGUU--UGGGAUGCGCUGAGGAGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCAGGAGCAAACGAGUUAAGUGC ((((.....((((.((((((.......)))--))...(((.((..((.(((((((.((((((..(((...))))))))).))))))))).)).)))..).))))..)))) ( -42.20, z-score = -1.74, R) >droSim1.chr2R 12654507 110 + 19596830 GCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGCUGGGGUCCUUGGGCUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGC .((((((....(..(....)..).....))))))...(..(((((((((((((((.((((((..(((...))))))))).))))))))))))).))..)........... ( -47.50, z-score = -2.14, R) >droSec1.super_1 11409439 110 + 14215200 GCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGAUGGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGC .((((((....(..(....)..).....))))))...(..(((..((((((((((.((((((..(((...))))))))).))))))))))...)))..)........... ( -43.10, z-score = -1.82, R) >consensus GCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGAUGGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGC (((((........))).........................((...(((((((((.((((((..(((...))))))))).))))))))).)).))............... (-36.60 = -36.27 + -0.33)


| Location | 13,915,816 – 13,915,924 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Shannon entropy | 0.11687 |
| G+C content | 0.62795 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -32.69 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13915816 108 - 21146708 GCACUUAACUCGUUUGCUCCUGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCUCCUCAGCGCAUCCCA--AACCCAUCCUGCCUGCCAGCUCCUUGGCGC ((.............(((...(((((((((.(((((((((...)))..)))))).)))))))))...)))........--..............(((((....))))))) ( -36.20, z-score = -2.56, R) >droSim1.chr2R 12654507 110 - 19596830 GCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGCCCAAGGACCCCAGCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGC ((.........(..((.(((.(((((((((.(.(((((((...)))..)))).).))))))))).)))))..).....................(((........))))) ( -35.70, z-score = -2.02, R) >droSec1.super_1 11409439 110 - 14215200 GCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCCAUCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGC ((.........(..((.((..(((((((((.(((((((((...)))..)))))).)))))))))..))))..).....................(((........))))) ( -36.20, z-score = -2.97, R) >consensus GCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCCAUCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGC ((...................(((((((((.(((((((((...)))..)))))).)))))))))..............................(((........))))) (-32.69 = -32.80 + 0.11)


| Location | 13,915,828 – 13,915,940 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.13473 |
| G+C content | 0.56756 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -34.86 |
| Energy contribution | -34.53 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13915828 112 + 21146708 UGGCAGGCAGGAUGGGUU--UGGGAUGCGCUGAGGAGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCAGGAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAG ......(((.......((--(((..((((((..((.(((((((.((((((..(((...))))))))).)))))))))...(((......))).))))))..)))))...))).. ( -42.20, z-score = -2.38, R) >droSim1.chr2R 12654519 114 + 19596830 AGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGCUGGGGUCCUUGGGCUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAA ...(((.(......).))).((((((..(((((((((((((((.((((((..(((...))))))))).))))))))))))).))..)..((.....)).))))).......... ( -45.70, z-score = -2.13, R) >droSec1.super_1 11409451 114 + 14215200 AGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGAUGGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAUAUAUGUAC ...(((.(......).))).((((((..(((..((((((((((.((((((..(((...))))))))).))))))))))...)))..)..((.....)).))))).......... ( -41.30, z-score = -1.57, R) >consensus AGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGAUGGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAA ..(((........((((............((...(((((((((.((((((..(((...))))))))).))))))))).)).(((...........))))))).......))).. (-34.86 = -34.53 + -0.33)


| Location | 13,915,828 – 13,915,940 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.13473 |
| G+C content | 0.56756 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -31.05 |
| Energy contribution | -31.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13915828 112 - 21146708 CUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUCCUGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCUCCUCAGCGCAUCCCA--AACCCAUCCUGCCUGCCA ............(((((.((.......)).)))))..(((((((((.(((((((((...)))..)))))).)))))))))..(((.(((.....--.........))))))... ( -35.44, z-score = -3.25, R) >droSim1.chr2R 12654519 114 - 19596830 UUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGCCCAAGGACCCCAGCACAUCCCCCCACCAACCCAUCCUGCCUGCCU ..........(((.(((((...........)))(((.(((((((((.(.(((((((...)))..)))).).))))))))).)))......................)))))... ( -32.30, z-score = -2.15, R) >droSec1.super_1 11409451 114 - 14215200 GUACAUAUAUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCCAUCACAUCCCCCCACCAACCCAUCCUGCCUGCCU ..........(((.(((((...((....))...))).(((((((((.(((((((((...)))..)))))).)))))))))..........................)))))... ( -34.20, z-score = -3.03, R) >consensus CUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCCAUCACAUCCCCCCACCAACCCAUCCUGCCUGCCU ..........(((.(((((...........)))....(((((((((.(((((((((...)))..)))))).)))))))))..........................)))))... (-31.05 = -31.17 + 0.11)

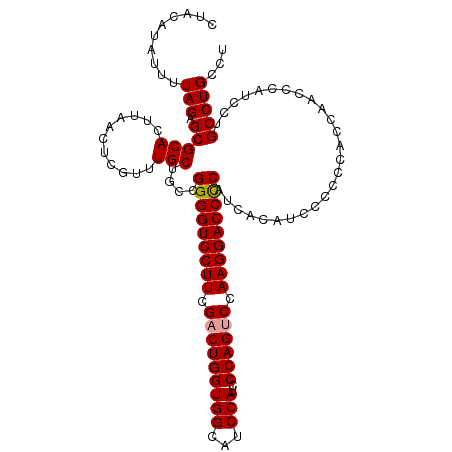

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:17 2011