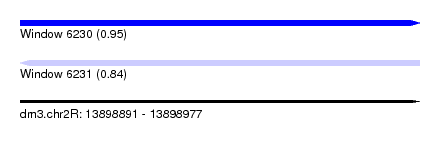
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,898,891 – 13,898,977 |
| Length | 86 |
| Max. P | 0.952140 |

| Location | 13,898,891 – 13,898,977 |
|---|---|
| Length | 86 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Shannon entropy | 0.45334 |
| G+C content | 0.34932 |
| Mean single sequence MFE | -13.61 |
| Consensus MFE | -7.89 |
| Energy contribution | -7.72 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13898891 86 + 21146708 GACAGCGAAAAUUUCAUUUCAUCUCACUGCAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUUUC----------GCAUCUACCUUUUAGCC ....(((((((................(((....((((((((((...)))))))))).--..))).)))))----------))............... ( -15.73, z-score = -2.90, R) >droAna3.scaffold_13266 9596378 86 - 19884421 GACAGCGAAAAUUUCAUUUCGUUUCACUGCAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUUUC----------ACUCCUACCUUAUUCAC ((.(((((((......))))))))).........((((((((((...)))))))))).--.((((......----------.....))))........ ( -15.80, z-score = -3.66, R) >droEre2.scaffold_4845 8127028 86 + 22589142 GACAGCGAAAAUUUCAUUUCAUUUCACUGCAUUUUACGUGAAAUUCAAUUUAACGUGC--UGGUAUUUCGC----------CCGUCCACCUUUUACCC (((.((((((..........(((((((..........)))))))........((....--..)).))))))----------..)))............ ( -13.00, z-score = -1.22, R) >droYak2.chr2R 5858711 96 - 21139217 GACAGCGAAAAUUUCAUUUAAUUUCACUGCAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUUUUUUCCCAACUACCAUCUACCUUUUACCC (.(((.((((...........)))).))))....((((((((((...)))))))))).--(((((.((........)).))))).............. ( -14.90, z-score = -3.63, R) >droSec1.super_1 11393232 86 + 14215200 GACAGCGAAAAUUUCAUUUCAUCUCACUGUAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUUUC----------UCAUCUACCUUUUAGCC ..((((((((......))))...............(((((((((...)))))))))))--)).........----------................. ( -13.80, z-score = -2.81, R) >droSim1.chr2R 12637155 86 + 19596830 GACAGCGAAAAUUUCAUUUCAUCUCACUGCAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUUUC----------GCAUCUACCUUUUAGCC ....(((((((................(((....((((((((((...)))))))))).--..))).)))))----------))............... ( -15.73, z-score = -2.90, R) >droPer1.super_2 6580690 86 + 9036312 GACAGGAAAAAUUUCAUUUCGUUUGGCUGAAUUUCACGUUAAAUUCAAUUUAACGUGCGCUUGUACUUCUC----------ACUUCCACCUAGUCU-- ((((((...........((((......))))...((((((((((...))))))))))..............----------.......))).))).-- ( -14.70, z-score = -1.27, R) >dp4.chr3 6364344 86 + 19779522 GACAGGAAAAAUUUCAUUUCGUUUCGCUGAAUUUCACGUUAAAUUCAAUUUAACGUGCGCUUGUACUUCUC----------ACUUCCACGUAGUCU-- .(((((...........((((......))))...((((((((((...))))))))))..))))).......----------...............-- ( -12.50, z-score = -0.35, R) >droVir3.scaffold_12875 5387671 80 - 20611582 GACAGCGAAAAUUUCAUUACGUUUCAUUGCAUUUCACGUUAAAUUCAAUUUAACGUGC--UGGUAUUUU-UC---------AUUUUUUAGCC------ ......(((((..(((.((((((..((((.((((......)))).))))..)))))).--)))...)))-))---------...........------ ( -14.00, z-score = -2.22, R) >droMoj3.scaffold_6496 8597879 73 + 26866924 GACAGCGAAAAUUUCAUUACGUUUCAUUGCAUUUCAAGUUAAAUUGAAUUUAACGUGC--UGGUAUUUUUUACGU----------------------- ..((((((.....))...(((((.........(((((......)))))...)))))))--))(((.....)))..----------------------- ( -10.70, z-score = -0.18, R) >droGri2.scaffold_15245 8114971 86 + 18325388 GACGGCGAAAAUUUCAUUACAUUUCAUUGCAUUUCACGUUAAAUUCAAUUCAACGUGC--UGGUAUUUUCUC---------ACUUUUUCCUCUUACC- ...((.((((((.(((.(((.((..((((.((((......)))).))))..)).))).--))).))))))))---------................- ( -8.90, z-score = -0.66, R) >consensus GACAGCGAAAAUUUCAUUUCAUUUCACUGCAUUUCACGUUAAAUUCAAUUUAACGUGC__UGGUAUUUUUC__________ACUUCUACCUUUUACCC .............................((...((((((((((...))))))))))...)).................................... ( -7.89 = -7.72 + -0.17)

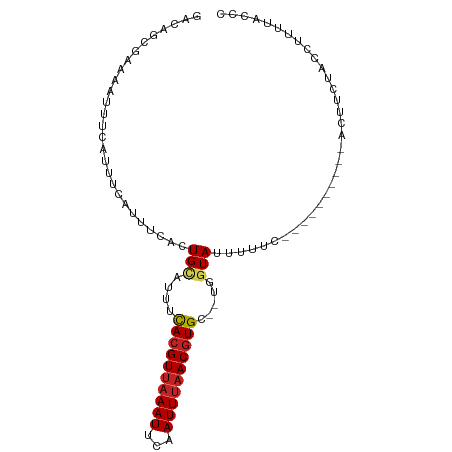
| Location | 13,898,891 – 13,898,977 |
|---|---|
| Length | 86 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Shannon entropy | 0.45334 |
| G+C content | 0.34932 |
| Mean single sequence MFE | -17.66 |
| Consensus MFE | -8.86 |
| Energy contribution | -9.25 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13898891 86 - 21146708 GGCUAAAAGGUAGAUGC----------GAAAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAGAUGAAAUGAAAUUUUCGCUGUC ........((((.....----------......)))).--.((((((((((...))))))))))....(((((((((...........))))))))). ( -21.50, z-score = -3.08, R) >droAna3.scaffold_13266 9596378 86 + 19884421 GUGAAUAAGGUAGGAGU----------GAAAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAAACGAAAUGAAAUUUUCGCUGUC ........((((....(----------....).)))).--.((((((((((...))))))))))....(((((((((...........))))))))). ( -20.80, z-score = -3.27, R) >droEre2.scaffold_4845 8127028 86 - 22589142 GGGUAAAAGGUGGACGG----------GCGAAAUACCA--GCACGUUAAAUUGAAUUUCACGUAAAAUGCAGUGAAAUGAAAUGAAAUUUUCGCUGUC ........((((((((.----------((.........--)).))))..(((..(((((((((.....)).)))))))..)))........))))... ( -16.00, z-score = -0.23, R) >droYak2.chr2R 5858711 96 + 21139217 GGGUAAAAGGUAGAUGGUAGUUGGGAAAAAAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAAAUUAAAUGAAAUUUUCGCUGUC ..............(((((.((........)).)))))--.((((((((((...))))))))))....(((((((((...........))))))))). ( -22.70, z-score = -3.61, R) >droSec1.super_1 11393232 86 - 14215200 GGCUAAAAGGUAGAUGA----------GAAAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUACAGUGAGAUGAAAUGAAAUUUUCGCUGUC ........((((.....----------......)))).--.((((((((((...))))))))))....(((((((((...........))))))))). ( -21.20, z-score = -3.92, R) >droSim1.chr2R 12637155 86 - 19596830 GGCUAAAAGGUAGAUGC----------GAAAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAGAUGAAAUGAAAUUUUCGCUGUC ........((((.....----------......)))).--.((((((((((...))))))))))....(((((((((...........))))))))). ( -21.50, z-score = -3.08, R) >droPer1.super_2 6580690 86 - 9036312 --AGACUAGGUGGAAGU----------GAGAAGUACAAGCGCACGUUAAAUUGAAUUUAACGUGAAAUUCAGCCAAACGAAAUGAAAUUUUUCCUGUC --.(((..(((.(((((----------(.....))).....((((((((((...))))))))))...))).)))....((((.......))))..))) ( -16.70, z-score = -0.77, R) >dp4.chr3 6364344 86 - 19779522 --AGACUACGUGGAAGU----------GAGAAGUACAAGCGCACGUUAAAUUGAAUUUAACGUGAAAUUCAGCGAAACGAAAUGAAAUUUUUCCUGUC --............((.----------(((((((.......((((((((((...))))))))))...((((.((...))...)))))))))))))... ( -15.80, z-score = -0.13, R) >droVir3.scaffold_12875 5387671 80 + 20611582 ------GGCUAAAAAAU---------GA-AAAAUACCA--GCACGUUAAAUUGAAUUUAACGUGAAAUGCAAUGAAACGUAAUGAAAUUUUCGCUGUC ------(((.((((...---------..-.........--.((((((((((...))))))))))...(((........)))......)))).)))... ( -12.40, z-score = -0.94, R) >droMoj3.scaffold_6496 8597879 73 - 26866924 -----------------------ACGUAAAAAAUACCA--GCACGUUAAAUUCAAUUUAACUUGAAAUGCAAUGAAACGUAAUGAAAUUUUCGCUGUC -----------------------.............((--(((((((..(((((.(((.....))).)).)))..)))))...((.....)))))).. ( -9.30, z-score = -0.32, R) >droGri2.scaffold_15245 8114971 86 - 18325388 -GGUAAGAGGAAAAAGU---------GAGAAAAUACCA--GCACGUUGAAUUGAAUUUAACGUGAAAUGCAAUGAAAUGUAAUGAAAUUUUCGCCGUC -..............((---------(((((.......--.((((((((((...))))))))))...((((......))))......))))))).... ( -16.40, z-score = -1.88, R) >consensus GGGUAAAAGGUAGAAGU__________GAAAAAUACCA__GCACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAAAUGAAAUGAAAUUUUCGCUGUC .........................................((((((((((...))))))))))....(((.(((((...........))))).))). ( -8.86 = -9.25 + 0.40)
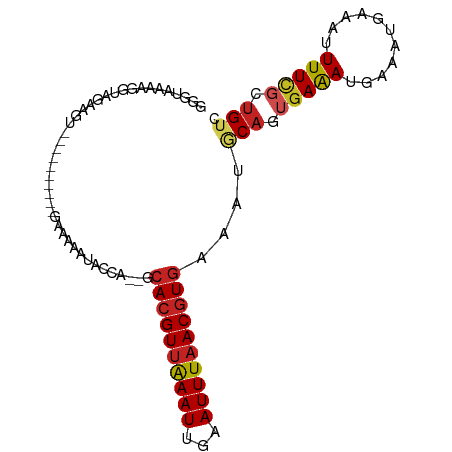
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:14 2011