
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,890,645 – 13,890,761 |
| Length | 116 |
| Max. P | 0.979915 |

| Location | 13,890,645 – 13,890,761 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Shannon entropy | 0.54630 |
| G+C content | 0.54897 |
| Mean single sequence MFE | -38.61 |
| Consensus MFE | -19.75 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
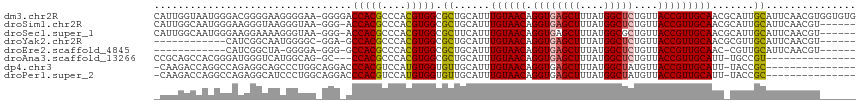
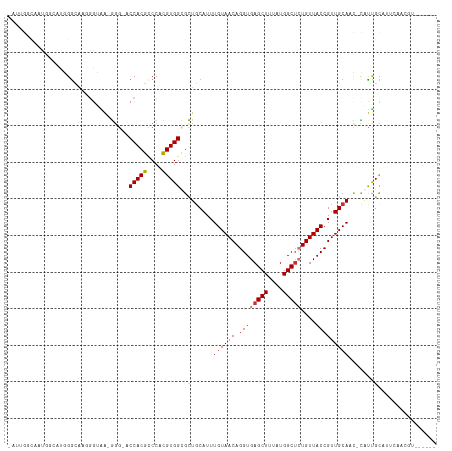
>dm3.chr2R 13890645 116 + 21146708 CAUUGGUAAUGGGACGGGGAAGGGGAA-GGGGACCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAACGCAUUGCAUUCAACGUGGUGUG .....((((((((((((((..((((..-(....)..).))).(((((.(((.((((((...))))))))).))))).))))))).)))))))..(((((..(.......)..))))) ( -41.20, z-score = -1.09, R) >droSim1.chr2R 12628446 109 + 19596830 CAUUGGCAAUGGGAAGGGUAAGGGUAA-GGG-ACCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAACGCAUUGCAUUCAACGU------ .....((((((.(....((((.(((((-.((-((((.(((.....)))(((.((((((...)))))))))....))).))).))))).))))..).)))))).........------ ( -39.10, z-score = -1.51, R) >droSec1.super_1 11385061 109 + 14215200 CAUUGGCAAUGGGAAGGAAAAGGGUAA-GGG-ACCACGCCCACGUGGCGCUUCAUUUGUAACAGGUGAGCUUUAUGGCGCUGUUACCGUUGCAACGCAUUGCAUUCAACGU------ .....((((((.(..........(.((-(.(-.(((((....)))))).))))..(((((((.((((((((....)))....))))))))))))).)))))).........------ ( -32.90, z-score = -0.09, R) >droYak2.chr2R 5850551 97 - 21139217 ------------CAUCGGCAAUGGGGC-GGA-GCCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAACGCGUUGCAUUCAACGU------ ------------.....((((((((((-(((-((((.(((.....)))(((.((((((...)))))))))....)))))))))).)))))))...((((((....))))))------ ( -42.70, z-score = -2.75, R) >droEre2.scaffold_4845 8119211 95 + 22589142 ------------CAUCGGCUA-GGGGA-GGG-GCCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAAC-CGUUGCAUUCAACGU------ ------------.....((.(-.((..-((.-((((((....)))))).))(((...((((((...(((((....)))))))))))...)))..)-).).)).........------ ( -34.10, z-score = -0.66, R) >droAna3.scaffold_13266 9589025 97 - 19884421 CCGCAGCCACGGGAUGGGUCAUGGCAG-GC---CCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAUU-UGCCGU--------------- ..((.((((((((.((((((......)-))---)))...)).)))))))).(((..((((((.((((((((....)))))....)))))))))..-)))...--------------- ( -44.70, z-score = -2.36, R) >dp4.chr3 6356039 100 + 19779522 -CAAGACCAGGCCAGAGGCAGCCCUGGCAGGACCCACGUCCAUGUGGUGUUGCAUUUGUAACAGGUGAGCUUUAUGGCUAUGUUACCGUUGCAUU-UACCGC--------------- -.........(((((.((...))))))).((((....))))..((((((.((((...((((((....((((....)))).))))))...))))..-))))))--------------- ( -37.10, z-score = -1.87, R) >droPer1.super_2 6572448 100 + 9036312 -CAAGACCAGGCCAGAGGCAUCCCUGGCAGGACCCACGUCCAUGUGGUGUUGCAUUUGUAACAGGUGAGCUUUAUGGCUAUGUUACCGUUGCAUU-UACCGC--------------- -.........(((((.((...))))))).((((....))))..((((((.((((...((((((....((((....)))).))))))...))))..-))))))--------------- ( -37.10, z-score = -2.16, R) >consensus _AUUGGCAAUGGCAUGGGCAAGGGUAA_GGG_ACCACGCCCACGUGGCGCUGCAUUUGUAACAGGUGAGCUUUAUGGCUCUGUUACCGUUGCAAC_CAUUGCAUUCAACGU______ .................................(((((....)))))...((((...(((((((...((((....)))))))))))...))))........................ (-19.75 = -20.06 + 0.31)

| Location | 13,890,645 – 13,890,761 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Shannon entropy | 0.54630 |
| G+C content | 0.54897 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
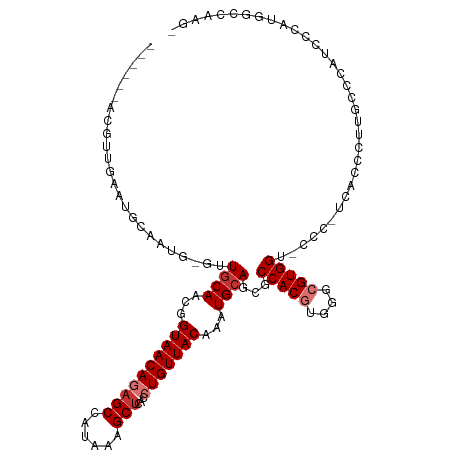
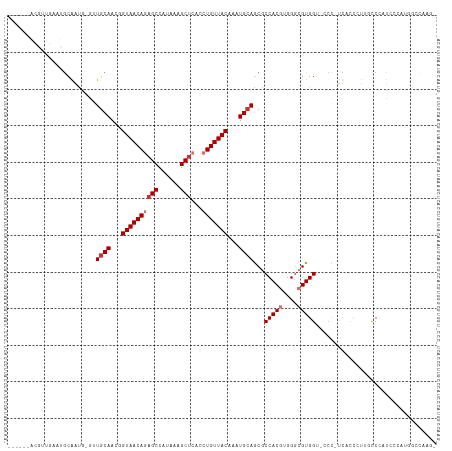
>dm3.chr2R 13890645 116 - 21146708 CACACCACGUUGAAUGCAAUGCGUUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGGUCCCC-UUCCCCUUCCCCGUCCCAUUACCAAUG .......(((((........((((((((...((((((((((......))))...))))))...))))))))...(((((((.((.....-........)).)).)))))...))))) ( -36.82, z-score = -3.12, R) >droSim1.chr2R 12628446 109 - 19596830 ------ACGUUGAAUGCAAUGCGUUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGGU-CCC-UUACCCUUACCCUUCCCAUUGCCAAUG ------.(((((...((((((.((((((...((((((((((......))))...))))))...))))))((((((....))))))-...-................))))))))))) ( -37.40, z-score = -3.18, R) >droSec1.super_1 11385061 109 - 14215200 ------ACGUUGAAUGCAAUGCGUUGCAACGGUAACAGCGCCAUAAAGCUCACCUGUUACAAAUGAAGCGCCACGUGGGCGUGGU-CCC-UUACCCUUUUCCUUCCCAUUGCCAAUG ------.(((((...((((((.(((.((...(((((((.((......))....)))))))...)).)))((((((....))))))-...-................))))))))))) ( -28.80, z-score = -0.59, R) >droYak2.chr2R 5850551 97 + 21139217 ------ACGUUGAAUGCAACGCGUUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGGC-UCC-GCCCCAUUGCCGAUG------------ ------.(((((...(((..((((((((...((((((((((......))))...))))))...))))))))...(((((.(((..-..)-)))))))))))))))------------ ( -39.40, z-score = -2.89, R) >droEre2.scaffold_4845 8119211 95 - 22589142 ------ACGUUGAAUGCAACG-GUUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGGC-CCC-UCCCC-UAGCCGAUG------------ ------.(((((....)))))-((((((...((((((((((......))))...))))))...))))))((((((....))))))-...-.....-.........------------ ( -35.50, z-score = -2.46, R) >droAna3.scaffold_13266 9589025 97 + 19884421 ---------------ACGGCA-AAUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGG---GC-CUGCCAUGACCCAUCCCGUGGCUGCGG ---------------.(.(((-..((((...((((((((((......))))...))))))...))))..(((((((((((((((---..-...))))).))))...))))))))).) ( -41.80, z-score = -2.30, R) >dp4.chr3 6356039 100 - 19779522 ---------------GCGGUA-AAUGCAACGGUAACAUAGCCAUAAAGCUCACCUGUUACAAAUGCAACACCACAUGGACGUGGGUCCUGCCAGGGCUGCCUCUGGCCUGGUCUUG- ---------------(.(((.-..((((...((((((.(((......)))....))))))...))))..))).)..((((....)))).((((((.(.......).))))))....- ( -31.40, z-score = -0.36, R) >droPer1.super_2 6572448 100 - 9036312 ---------------GCGGUA-AAUGCAACGGUAACAUAGCCAUAAAGCUCACCUGUUACAAAUGCAACACCACAUGGACGUGGGUCCUGCCAGGGAUGCCUCUGGCCUGGUCUUG- ---------------......-..((((...((((((.(((......)))....))))))...))))...((((......))))(.((.(((((((....)))))))..)).)...- ( -31.50, z-score = -0.64, R) >consensus ______ACGUUGAAUGCAAUG_GUUGCAACGGUAACAGAGCCAUAAAGCUCACCUGUUACAAAUGCAGCGCCACGUGGGCGUGGU_CCC_UCACCCUUGCCCAUCCCAUGGCCAAG_ ........................((((...((((((((((......)))...)))))))...))))...(((((....)))))................................. (-18.76 = -19.51 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:13 2011