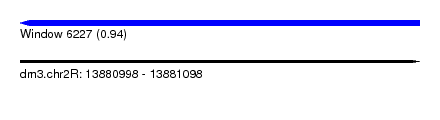
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,880,998 – 13,881,098 |
| Length | 100 |
| Max. P | 0.938576 |

| Location | 13,880,998 – 13,881,098 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.85 |
| Shannon entropy | 0.56461 |
| G+C content | 0.46264 |
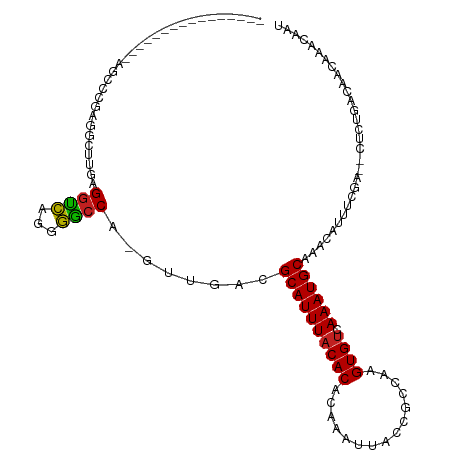
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -12.94 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13880998 100 - 21146708 -------------CACAGCCCGAGGCUUGAGGUCAGGGGCCACAUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGA--CUCUGGCAACAAACAAU -------------....(((.(((..((((((((((.(....).)))))((((((((((...............)))).))))))......)))))--))).))).......... ( -24.66, z-score = -1.44, R) >droSim1.chr2R 12617178 100 - 19596830 -------------CACAGCCCGAGGCUUGAGGUCAGGGGCCACAUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGA--CUCUGGCAACAAACAAU -------------....(((.(((..((((((((((.(....).)))))((((((((((...............)))).))))))......)))))--))).))).......... ( -24.66, z-score = -1.44, R) >droSec1.super_1 11375385 102 - 14215200 -------------CACAGCUCGAGGCUUGAGGUCAGGGGCCACAUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGACUCUCUGGCAACAAACAAU -------------....(((.(((..((((((((((.(....).)))))((((((((((...............)))).))))))......)))))..))).))).......... ( -25.96, z-score = -2.07, R) >droYak2.chr2R 5840584 113 + 21139217 CCCGAGCUCGAACCGGAGCCCGAGGCUUGAGGUCAGGGGCCACAUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGA--CACUGGCAACAAACAAU (((((.(((((.((((...))).)..))))).)).)))...............................((((.((((((((((....))))).))--))))))).......... ( -32.00, z-score = -2.01, R) >droEre2.scaffold_4845 8109758 97 - 22589142 ----------------AGCCCCAGGCUUGAGGUCAGGGGCCACAUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGA--CUCUGGCAACAAACAAU ----------------.(((((.(((.....))).))))).............................((((.(.((((((((....))))).))--).))))).......... ( -26.20, z-score = -2.04, R) >droAna3.scaffold_13266 9579275 104 + 19884421 --------AGGAAGAAGCCCUCGGGCUUGAGGUCAGGGGCCA-GUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUCUGA--CUGUAACAACAAACAAU --------......(((((....)))))..((((((((....-(((...((((((((((...............)))).)))))).))).))))))--))............... ( -25.86, z-score = -0.81, R) >dp4.chr3 6347099 102 - 19779522 -----------GAGGGGCAGGGACUCUCCUGGGGAGCCUCCA-GUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUUGA-AAACAGACAACAAACAAU -----------(.(((((.(((.....))).....)))))).-((((..((((((((((...............)))).))))))......((((.-...))))))))....... ( -22.56, z-score = -0.46, R) >droPer1.super_2 6562999 102 - 9036312 -----------GAGGGGCAGGGGCUCUCCUGGGGAGCCUCCA-GUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUUGA-AAACAGACAACAAACAAU -----------........((((((((.....)))))).)).-((((..((((((((((...............)))).))))))......((((.-...))))))))....... ( -26.86, z-score = -1.41, R) >droWil1.scaffold_180745 2408056 76 - 2843958 ---------------------------UAUGGCCAGAGGCCA-GUUGACGCAUUUACACA-AAAUUACCGCCAAGUGUCAAAUGCAAACAUUU-AG--CUGUAACAAU------- ---------------------------..(((((...)))))-((((..((((((((((.-.............)))).))))))..(((...-..--.)))..))))------- ( -16.94, z-score = -1.41, R) >droVir3.scaffold_12875 5362474 77 + 20611582 --------------------------UCAUGGCCUGGGGCCA-GUUGACGCAUUUACACA-AAAUUACCGCCAAGUGUCAAAUGCAAACAUUU-UG--UUUCAACAAU------- --------------------------...(((((...)))))-(((((.((((((((((.-.............)))).)))))).(((....-.)--)))))))...------- ( -20.34, z-score = -1.80, R) >droMoj3.scaffold_6496 8565154 88 - 26866924 ---------------UUCGGUUCGCUUCCCGCCCGGCGGCCA-GUUGACGCAUUUUCACU-AAAUUACCGCCAAGUGUCAAAUGCAAACAUUU-CG--CUUCAACAAU------- ---------------...(((.((((........))))))).-(((((.((.....((((-............))))..(((((....)))))-.)--).)))))...------- ( -18.40, z-score = -0.56, R) >droGri2.scaffold_15245 8093931 91 - 18325388 -------------UUUUUGGGCCAGAUCAUGGCCAGAGGCCA-GUUGUCGCAUUUACACA-AAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGG--CUUUAACAAU------- -------------...((((((((.....))))).((((((.-..(((.((((((((((.-.............)))).))))))..)))....))--))))..))).------- ( -26.54, z-score = -2.74, R) >consensus ________________AGCCCGAGGCUUGAGGUCAGGGGCCA_GUUGACGCAUUUACACACAAAUUACCGCCAAGUGUCAAAUGCAAACAUUUCGA__CUCUGACAACAAACAAU ..............................((((...))))........((((((((((...............)))).)))))).............................. (-12.94 = -12.38 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:11 2011