
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,879,387 – 13,879,443 |
| Length | 56 |
| Max. P | 0.732106 |

| Location | 13,879,387 – 13,879,443 |
|---|---|
| Length | 56 |
| Sequences | 11 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Shannon entropy | 0.31896 |
| G+C content | 0.52528 |
| Mean single sequence MFE | -18.29 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
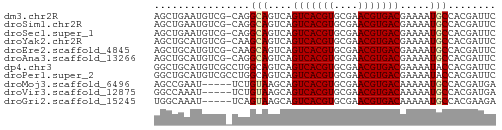
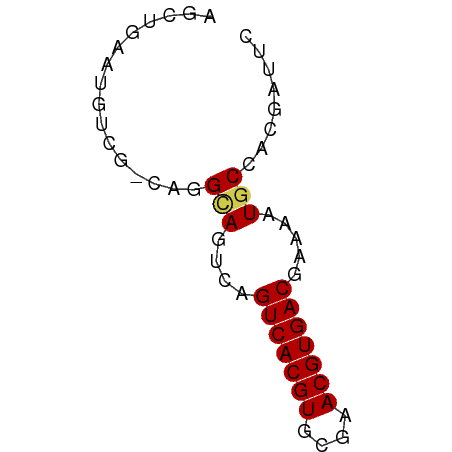
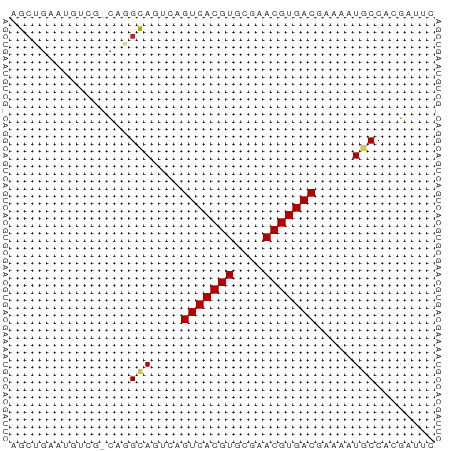
>dm3.chr2R 13879387 56 - 21146708 AGCUGAAUGUCG-CAGGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-..((((.((.(((((((....)))))))))...)))).)))).. ( -21.10, z-score = -2.42, R) >droSim1.chr2R 12615543 56 - 19596830 AGCUGAAUGUCG-CAGGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-..((((.((.(((((((....)))))))))...)))).)))).. ( -21.10, z-score = -2.42, R) >droSec1.super_1 11373747 56 - 14215200 AGCUGAAUGUCG-CAGGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-..((((.((.(((((((....)))))))))...)))).)))).. ( -21.10, z-score = -2.42, R) >droYak2.chr2R 5838927 56 + 21139217 AGCUGCAUGUCG-CAAGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-...(((.((.(((((((....)))))))))...)))..)))).. ( -17.60, z-score = -1.00, R) >droEre2.scaffold_4845 8108199 56 - 22589142 AGCUGCAUGUCG-CAAGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-...(((.((.(((((((....)))))))))...)))..)))).. ( -17.60, z-score = -1.00, R) >droAna3.scaffold_13266 9577531 56 + 19884421 AGCUGCAUGUCG-CAGGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ........((((-..((((.((.(((((((....)))))))))...)))).)))).. ( -21.10, z-score = -2.00, R) >dp4.chr3 6345212 57 - 19779522 GGCUGCAUGUCGCCUGGCAGUCAGUCACGUGCGAACGUGACGAAAAUACCACGAUUC ((((((..........)))))).(((((((....)))))))................ ( -19.40, z-score = -1.56, R) >droPer1.super_2 6561105 57 - 9036312 GGCUGCAUGUCGCCUGGCAGUCAGUCACGUGCGAACGUGACGAAAAUACCACGAUUC ((((((..........)))))).(((((((....)))))))................ ( -19.40, z-score = -1.56, R) >droMoj3.scaffold_6496 8561806 52 - 26866924 AGCCGAAU-----UCUGUAAGCAGUCACGUGCGAACGUGACAAAAAUGCCACGAUGA ........-----.......((((((((((....))))))).....)))........ ( -11.30, z-score = -0.53, R) >droVir3.scaffold_12875 5360106 52 + 20611582 GGCCAAAU-----UCUGUAAGCAGUCACGUGCGAACGUGACAAAAAUGCCACGAUGA (((.....-----..((....))(((((((....)))))))......)))....... ( -14.90, z-score = -1.98, R) >droGri2.scaffold_15245 8091389 52 - 18325388 UGGCAAAU-----UCAGUAAGCAGUCACGUGCGAACGUGACAAAAAUGCCACGAAGA (((((...-----...(....).(((((((....))))))).....)))))...... ( -16.60, z-score = -3.09, R) >consensus AGCUGAAUGUCG_CAGGCAGUCAGUCACGUGCGAACGUGACGAAAAUGCCACGAUUC ................(((....(((((((....))))))).....)))........ (-12.08 = -12.11 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:10 2011