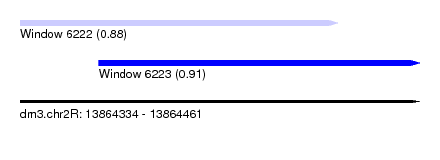
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,864,334 – 13,864,461 |
| Length | 127 |
| Max. P | 0.910344 |

| Location | 13,864,334 – 13,864,435 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Shannon entropy | 0.30509 |
| G+C content | 0.45420 |
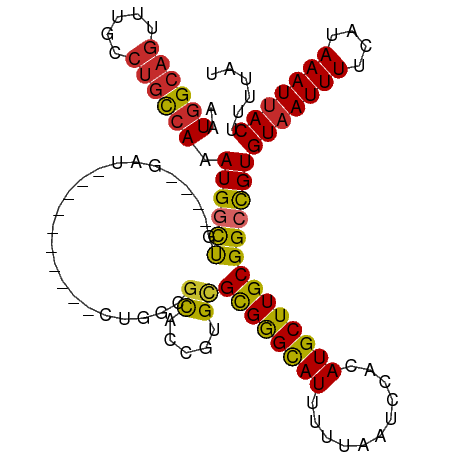
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
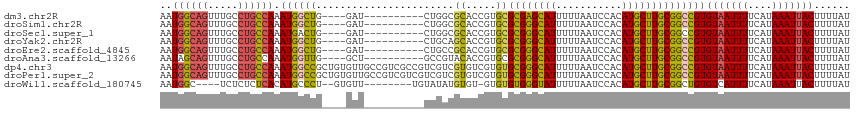
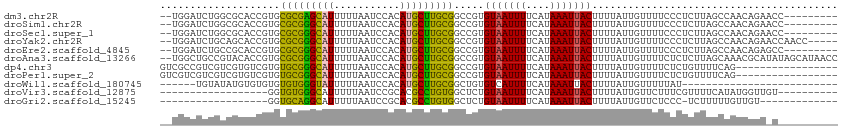
>dm3.chr2R 13864334 101 + 21146708 AAUGGCAGUUUGCCUGCCAAAUGGCUG----GAU----------CUGGCGCACCGUGCGCGAGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((.----...----------...((((...))))((((((((...........))))))))))))))(((((((....)))))))...... ( -34.30, z-score = -2.26, R) >droSim1.chr2R 12600185 101 + 19596830 AAUGGCAGUUUGCCUGCCAAAUGGCUG----GAU----------CUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((.----...----------...((((...))))((((((((...........))))))))))))))(((((((....)))))))...... ( -33.50, z-score = -1.52, R) >droSec1.super_1 11358846 101 + 14215200 AAUGGCAGUUUGCCUGCCAAAUGACUG----GAU----------CUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU .(((((.((.((((..(((......))----)..----------..)))).))....(((((((((...........))))))))))))))(((((((....)))))))...... ( -31.10, z-score = -1.38, R) >droYak2.chr2R 5823348 101 - 21139217 AAUGGCAGUUUGCCUGCCAAAUGGCUG----GAU----------CUGCAGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((.----...----------..(((......)))((((((((...........))))))))))))))(((((((....)))))))...... ( -32.20, z-score = -1.36, R) >droEre2.scaffold_4845 8092618 101 + 22589142 AAUGGCAGUUUGCCUGCCAAAUGGCUG----GAU----------CUGCCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ...(((((....((.(((....))).)----)..----------)))))((((.((.(((((((((...........))))))))))).))))...................... ( -34.20, z-score = -1.97, R) >droAna3.scaffold_13266 9563054 101 - 19884421 AAUAGCAGUUUGCCUGCCAAAUGGUUG----GCU----------GCCGUACACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ....(((((..(((........)))..----)))----------))..(((((.((.(((((((((...........))))))))))).)))))..................... ( -30.40, z-score = -1.25, R) >dp4.chr3 6330871 115 + 19779522 AAUGGCAGUUUGCCUGCCAAAUGGCCGCUGUGUUGCCGUCGCCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((((.......(((.(((((.(.....).)).))))))((((...........))))..))))))))(((((((....)))))))...... ( -38.50, z-score = -2.05, R) >droPer1.super_2 6546770 115 + 9036312 AAUGGCAGUUUGCCUGCCAAAUGGCCGCUGUGUUGCCGUCGUCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((((......((((.((....)).)).)).(((((.(((.........))).)))))..))))))))(((((((....)))))))...... ( -37.70, z-score = -2.21, R) >droWil1.scaffold_180745 2381962 100 + 2843958 AAUGGC----UCUCUCUCACAUGCCCU--GUGUU--------UGUAUAUGUGU-GUGUGUGGGUAUUUUUAAUCCACAUGCUUGCGGCUGUGUCAUUUUCAUAAAUUACUUUUAU .((((.----.......((((.(((..--((((.--------...))))(((.-((((((((((.......)))))))))).))))))))))......))))............. ( -22.24, z-score = -1.56, R) >consensus AAUGGCAGUUUGCCUGCCAAAUGGCUG____GAU__________CUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAU ..((((((.....)))))).((((((.......................((.....))((((((((...........))))))))))))))(((((((....)))))))...... (-25.35 = -25.02 + -0.33)
| Location | 13,864,359 – 13,864,461 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.58009 |
| G+C content | 0.41551 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -13.33 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
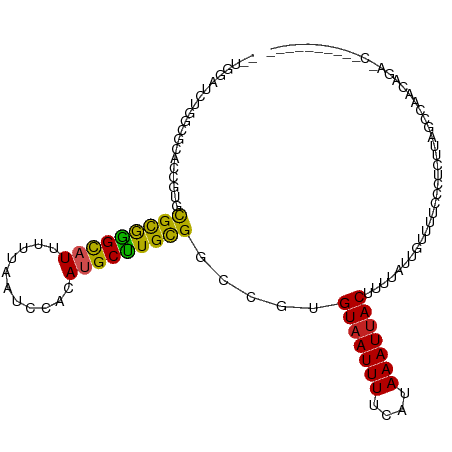
>dm3.chr2R 13864359 102 + 21146708 --UGGAUCUGGCGCACCGUGCGCGAGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGAACC--------- --.((.((((..((..((..(((((((((...........)))))))))..)).(((((((....)))))))....................))...)))).))--------- ( -25.50, z-score = -1.82, R) >droSim1.chr2R 12600210 102 + 19596830 --UGGAUCUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGAACC--------- --.((.((((..((..((..(((((((((...........)))))))))..)).(((((((....)))))))....................))...)))).))--------- ( -24.70, z-score = -0.84, R) >droSec1.super_1 11358871 102 + 14215200 --UGGAUCUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGAACC--------- --.((.((((..((..((..(((((((((...........)))))))))..)).(((((((....)))))))....................))...)))).))--------- ( -24.70, z-score = -0.84, R) >droYak2.chr2R 5823373 106 - 21139217 --UGGAUCUGCAGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGAACCAACC----- --(((.((((..((..((..(((((((((...........)))))))))..)).(((((((....)))))))....................))...)))).)))...----- ( -26.00, z-score = -1.64, R) >droEre2.scaffold_4845 8092643 102 + 22589142 --UGGAUCUGCCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGAGCC--------- --.(((...(((((((((....)))((((...........)))).))))))...(((((((....)))))))...........)))((((........))))..--------- ( -24.10, z-score = -0.68, R) >droAna3.scaffold_13266 9563079 111 - 19884421 --UGGCUGCCGUACACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUCUUAGCAAACGCAUAUAGCAUAACC --..((((((((((...))))((((((((...........)))))))))))((((((((((....))))))).....(((((.........)))))))).....)))...... ( -25.40, z-score = -0.81, R) >dp4.chr3 6330908 96 + 19779522 GUCGCCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUUCAG----------------- ...(((..((......))...((((((((...........)))))))))))...(((((((....)))))))..............(((....)))----------------- ( -17.50, z-score = -0.47, R) >droPer1.super_2 6546807 96 + 9036312 GUCGUCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUUCAG----------------- .........((((((.((((((.((..........))))))))..))))))...(((((((....)))))))..............(((....)))----------------- ( -16.10, z-score = -0.33, R) >droWil1.scaffold_180745 2381990 81 + 2843958 ------UGUAUAUGUGUGUGUGUGGGUAUUUUUAAUCCACAUGCUUGCGGCUGUGUCAUUUUCAUAAAUUACUUUUAUUGUUUUUAU-------------------------- ------.(((..((((.((((((((((.......)))))))))).))))..((((.......))))...)))...............-------------------------- ( -14.80, z-score = -0.86, R) >droVir3.scaffold_12875 5339176 85 - 20611582 ------------------GGUGUGGGCAUUUUUAAUCCGCACGCCUGUGGCUCUGUAAUUUUCAUAAAUUACUUUUAUUGUUCUUUCGUUUUCAUAUGGUUGU---------- ------------------.(((((((.........)))))))((((((((....(((((((....))))))).......(......)....))))).)))...---------- ( -15.50, z-score = -1.06, R) >droGri2.scaffold_15245 8072582 81 + 18325388 ------------------GGUGCAGGCAUUUUUAAUCCGCACGCCUGUGGCUCUGUAAUUUUCAUAAAUUACUUUUAUUGUUCUCCC-UCUUUUUGUUGU------------- ------------------(.(((((((...............))))))).)...(((((((....)))))))...............-............------------- ( -13.56, z-score = -1.08, R) >consensus __UGGAUCUGGCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAACAGA_C__________ ....................(((((((((...........))))))))).....(((((((....)))))))......................................... (-13.33 = -12.66 + -0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:08 2011