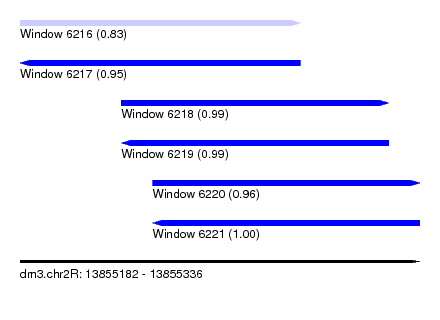
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,855,182 – 13,855,336 |
| Length | 154 |
| Max. P | 0.998698 |

| Location | 13,855,182 – 13,855,290 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Shannon entropy | 0.28403 |
| G+C content | 0.40306 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
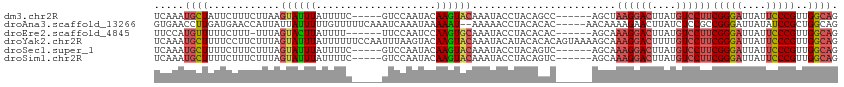
>dm3.chr2R 13855182 108 + 21146708 AAUAAAGAAAAACAAAA-GGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUAGCU------GGCUGUAGGUAUUUGU---- .......(((.((...(-(.((((.....(((........)))(((((((...((((..(((((....)))))..)).)))))))))...)))------).))....)).)))..---- ( -27.60, z-score = -2.14, R) >droAna3.scaffold_13266 9553551 112 - 19884421 AAGAAAGAAGACUGGGAAAGC--UGGGAUGAGAGGUUUAACUUGGAUAUAACCUGCCAGCGGAUAUAAUCCCGGCGGAGAUAAGUUCUUUUGUU-----GUGUGUAGGUUUUUAUUUUU ...........((.((....)--).))...((((((..((((((.((((((((((((.(.((((...))))))))))(((.....)))...)))-----)))).))))))...)))))) ( -25.50, z-score = -0.14, R) >droEre2.scaffold_4845 8083567 108 + 22589142 AACAAAGAAAAACAAAA-GGCAGCGGAAUGGGAUGAUAAACUUGGGUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GUGUGUAGGUAUUUGC---- ..((((.....(((...-.(((((.....(((((..(((((....))))).((((....))))....)))))(((((((....))))))))))------)).))).....)))).---- ( -32.20, z-score = -2.67, R) >droYak2.chr2R 5813869 114 - 21139217 AACAAAGAAAAACAAAA-GGCAGCGGCAGGGGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAAAAGUCCUUUGCUUUUACUGUGUGUAUGUAUUUGU---- .(((((.....((((((-((((..((((((....(((........)))...))))))..(((((....)))))((((((....)))))))))))))....))).......)))))---- ( -34.70, z-score = -3.05, R) >droSec1.super_1 11349786 109 + 14215200 AACAAAGAAAAACAAAAAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGU---- .(((((.....((....((.(((((((..(((........)))(((((((...((((..(((((....)))))..)).)))))))))))))))------).))....)).)))))---- ( -28.10, z-score = -2.67, R) >droSim1.chr2R 12591046 109 + 19596830 AACAAAGAAAAACAAAAAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGU---- .(((((.....((....((.(((((((..(((........)))(((((((...((((..(((((....)))))..)).)))))))))))))))------).))....)).)))))---- ( -28.10, z-score = -2.67, R) >consensus AACAAAGAAAAACAAAA_GGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU______GUCUGUAGGUAUUUGU____ ............((((.............(((........))).......((((((...(((((....)))))((((((....))))))..............)))))).))))..... (-18.69 = -18.33 + -0.36)


| Location | 13,855,182 – 13,855,290 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Shannon entropy | 0.28403 |
| G+C content | 0.40306 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13855182 108 - 21146708 ----ACAAAUACCUACAGCC------AGCUAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCC-UUUUGUUUUUCUUUAUU ----(((((........(((------(((.((((((....)))))).((((....))))))))))((((........((........)).........)))-))))))........... ( -27.23, z-score = -3.29, R) >droAna3.scaffold_13266 9553551 112 + 19884421 AAAAAUAAAAACCUACACAC-----AACAAAAGAACUUAUCUCCGCCGGGAUUAUAUCCGCUGGCAGGUUAUAUCCAAGUUAAACCUCUCAUCCCA--GCUUUCCCAGUCUUCUUUCUU ....................-----....((((((.............((((...))))(((((.(((((............)))))......)))--))..........))))))... ( -14.20, z-score = -0.30, R) >droEre2.scaffold_4845 8083567 108 - 22589142 ----GCAAAUACCUACACAC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAACCCAAGUUUAUCAUCCCAUUCCGCUGCC-UUUUGUUUUUCUUUGUU ----(((((.....(((..(------(((.((((((....))))))(((((....))))).(((...(.(((((....))))).)...)))....))))..-...))).....))))). ( -26.60, z-score = -2.59, R) >droYak2.chr2R 5813869 114 + 21139217 ----ACAAAUACAUACACACAGUAAAAGCAAAGGACUUUUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCCCCUGCCGCUGCC-UUUUGUUUUUCUUUGUU ----............(((.((.(((((((((((((....))))..(((((....)))))..((((((.(((((....)))))......))))))......-.))))))))))).))). ( -32.00, z-score = -3.78, R) >droSec1.super_1 11349786 109 - 14215200 ----ACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUUUUUGUUUUUCUUUGUU ----(((((.....((((((------(((.((((((....)))))).((((....))))))))))((((........((........)).........))))...))).....))))). ( -26.43, z-score = -3.02, R) >droSim1.chr2R 12591046 109 - 19596830 ----ACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUUUUUGUUUUUCUUUGUU ----(((((.....((((((------(((.((((((....)))))).((((....))))))))))((((........((........)).........))))...))).....))))). ( -26.43, z-score = -3.02, R) >consensus ____ACAAAUACCUACACAC______AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCC_UUUUGUUUUUCUUUGUU ..........................(((((((((....((((...(((((....)))))..))))((......))..................................))))))))) (-16.09 = -16.82 + 0.72)


| Location | 13,855,221 – 13,855,324 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Shannon entropy | 0.32632 |
| G+C content | 0.36848 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13855221 103 + 21146708 CUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUAGCU------GGCUGUAGGUAUUUGUACUUGU------AUUGGACGAAAAUAAAUACUUAAGAAAG ...((......)).((((.(((((....)))))((((((....))))))...)------)))..(((((((((((..((((------.....))))..)))))))))))...... ( -31.70, z-score = -3.89, R) >droAna3.scaffold_13266 9553589 107 - 19884421 CUUGGAUAUAACCUGCCAGCGGAUAUAAUCCCGGCGGAGAUAAGUUCUUUUGUU-----GUGUGUAGGUUUUUAUUUUUAU---UUGAUUUGAAAAACAAAAAUAAUAAUGGUUC ((((.((((((((((((.(.((((...))))))))))(((.....)))...)))-----)))).))))...(((((.((((---((...((....))...)))))).)))))... ( -22.40, z-score = -1.07, R) >droEre2.scaffold_4845 8083606 101 + 22589142 CUUGGGUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GUGUGUAGGUAUUUGCACUUG------GAUUGGA-AAAAAUAAGUACUAAAAAAG- .((((((...((((((((.(((((....)))))((((((....))))))....------.)).))))))....))(((((------..((...-.))..))))).)))).....- ( -28.90, z-score = -2.64, R) >droYak2.chr2R 5813908 114 - 21139217 CUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAAAAGUCCUUUGCUUUUACUGUGUGUAUGUAUUUGUACUUGUACUUAAAUUGGA-AAAAAUAAAUACUAAAGAAGG ...(((((...((((....))))...)))))((((((((....))))))))(((((...((((.(((((((........)))).....((...-.)).))).))))...))))). ( -24.70, z-score = -1.78, R) >droSec1.super_1 11349826 103 + 14215200 CUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGUACUUGU------AUUGGACGAAAAUAAAUACUAAAGAAAG ...(((((...((((....))))...)))))((((((((....))))))))..------..((...(((((((((..((((------.....))))..)))))))))..)).... ( -28.30, z-score = -3.47, R) >droSim1.chr2R 12591086 103 + 19596830 CUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGUACUUGU------AUUGGACGAAAAUAAAUACUAAAGAAAG ...(((((...((((....))))...)))))((((((((....))))))))..------..((...(((((((((..((((------.....))))..)))))))))..)).... ( -28.30, z-score = -3.47, R) >consensus CUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU______GUCUGUAGGUAUUUGUACUUGU______AUUGGACAAAAAUAAAUACUAAAGAAAG ..........((((((...(((((....)))))((((((....))))))..............)))))).............................................. (-18.23 = -18.78 + 0.56)


| Location | 13,855,221 – 13,855,324 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Shannon entropy | 0.32632 |
| G+C content | 0.36848 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13855221 103 - 21146708 CUUUCUUAAGUAUUUAUUUUCGUCCAAU------ACAAGUACAAAUACCUACAGCC------AGCUAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAG .........((((((.............------..))))))....((((...(((------(((.((((((....)))))).((((....)))))))))))))).......... ( -27.76, z-score = -4.03, R) >droAna3.scaffold_13266 9553589 107 + 19884421 GAACCAUUAUUAUUUUUGUUUUUCAAAUCAA---AUAAAAAUAAAAACCUACACAC-----AACAAAAGAACUUAUCUCCGCCGGGAUUAUAUCCGCUGGCAGGUUAUAUCCAAG .((((....((((((((((((........))---))))))))))............-----...................((((((........).))))).))))......... ( -18.10, z-score = -2.55, R) >droEre2.scaffold_4845 8083606 101 - 22589142 -CUUUUUUAGUACUUAUUUUU-UCCAAUC------CAAGUGCAAAUACCUACACAC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAACCCAAG -........((((((......-.......------.))))))....((((.....(------(((.((((((....)))))).((((....))))))))..)))).......... ( -24.44, z-score = -2.96, R) >droYak2.chr2R 5813908 114 + 21139217 CCUUCUUUAGUAUUUAUUUUU-UCCAAUUUAAGUACAAGUACAAAUACAUACACACAGUAAAAGCAAAGGACUUUUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAG (((......(((((((..((.-...))..)))))))...........................((.((((((....))))))(((((....)))))...)))))........... ( -22.90, z-score = -2.37, R) >droSec1.super_1 11349826 103 - 14215200 CUUUCUUUAGUAUUUAUUUUCGUCCAAU------ACAAGUACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAG .........((((((.............------..))))))....((((...(((------(((.((((((....)))))).((((....)))))))))))))).......... ( -25.66, z-score = -3.72, R) >droSim1.chr2R 12591086 103 - 19596830 CUUUCUUUAGUAUUUAUUUUCGUCCAAU------ACAAGUACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAG .........((((((.............------..))))))....((((...(((------(((.((((((....)))))).((((....)))))))))))))).......... ( -25.66, z-score = -3.72, R) >consensus CUUUCUUUAGUAUUUAUUUUCGUCCAAU______ACAAGUACAAAUACCUACACAC______AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAG .........((((((.....................))))))....((((.((.............((((((....))))))(((((....))))).))..)))).......... (-14.45 = -15.23 + 0.78)

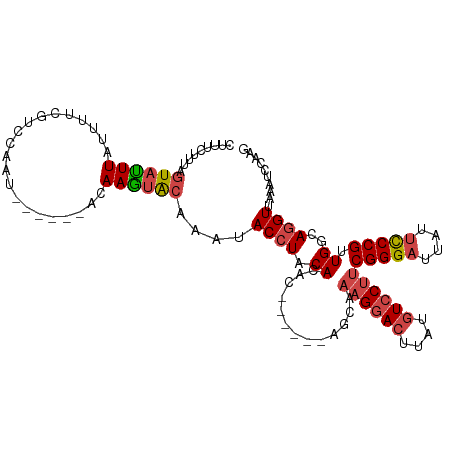

| Location | 13,855,233 – 13,855,336 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.38888 |
| G+C content | 0.36042 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -14.65 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13855233 103 + 21146708 CUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUAGCU------GGCUGUAGGUAUUUGUACUUGUAUUGGAC-----GAAAAUAAAUACUUAAGAAAGAAUAGCAUUUGA ..((((.(((((....)))))((((((....))))))...)------)))..(((((((((((..((((.....))-----))..))))))))))).................. ( -31.50, z-score = -4.03, R) >droAna3.scaffold_13266 9553601 107 - 19884421 CUGCCAGCGGAUAUAAUCCCGGCGGAGAUAAGUUCUUUUGUU-----GUGUGUAGGUUUUU--AUUUUUAUUUGAUUUGAAAAACAAAAAUAAUAAUGGUUCAUCAAGGUUCAC (((((.(.((((...)))))))))).(((..(..((.(((((-----((.(((...(((((--(..((.....))..)))))))))...))))))).))..))))......... ( -18.70, z-score = 0.20, R) >droEre2.scaffold_4845 8083618 101 + 22589142 CUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GUGUGUAGGUAUUUGCACUUGGAUUGGAA------AAAAUAAGUACUAAA-AAAGAAAAACAUGGAA .((((.((((((....))))(((((((....)))))))...------....)).)))).....(((((..((....------))..)))))......-................ ( -22.90, z-score = -1.63, R) >droYak2.chr2R 5813920 114 - 21139217 CUGCCAACGGGAAUAAUCCCGAAGGACAAAAGUCCUUUGCUUUUACUGUGUGUAUGUAUUUGUACUUGUACUUAAAUUGGAAAAAAUAAAUACUAAAGAAGGAAAAGCAUUUGA .......(((((....)))))((((((....))))))(((((((.((.....((.((((((((..((...(.......)..))..)))))))))).....)))))))))..... ( -26.10, z-score = -2.36, R) >droSec1.super_1 11349838 103 + 14215200 CUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGUACUUGUAUUGGAC-----GAAAAUAAAUACUAAAGAAAGAAAAGCAUUUGA .......(((((....)))))((((((....))))))((((------..((.(.(((((((((..((((.....))-----))..))))))))).)....))...))))..... ( -26.60, z-score = -3.20, R) >droSim1.chr2R 12591098 103 + 19596830 CUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU------GACUGUAGGUAUUUGUACUUGUAUUGGAC-----GAAAAUAAAUACUAAAGAAAGAAAAGCAUUUGA .......(((((....)))))((((((....))))))((((------..((.(.(((((((((..((((.....))-----))..))))))))).)....))...))))..... ( -26.60, z-score = -3.20, R) >consensus CUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCUUUGCU______GUCUGUAGGUAUUUGUACUUGUAUUGGAC_____GAAAAUAAAUACUAAAGAAAGAAAAGCAUUUGA .((((.((((((....))))(((((((....))))))).............)).))))........................................................ (-14.65 = -15.40 + 0.75)


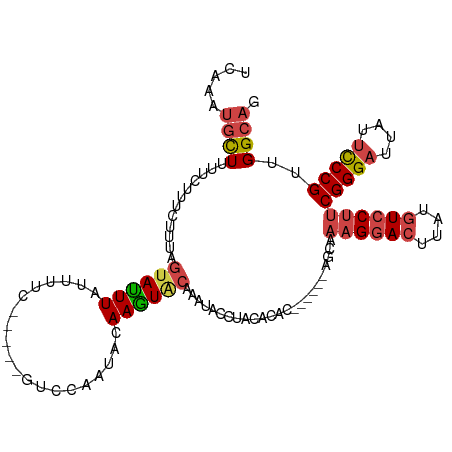
| Location | 13,855,233 – 13,855,336 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.38888 |
| G+C content | 0.36042 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.998698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13855233 103 - 21146708 UCAAAUGCUAUUCUUUCUUAAGUAUUUAUUUUC-----GUCCAAUACAAGUACAAAUACCUACAGCC------AGCUAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAG ..(((((((...........)))))))......-----..........................(((------(((.((((((....)))))).((((....)))))))))).. ( -27.20, z-score = -4.39, R) >droAna3.scaffold_13266 9553601 107 + 19884421 GUGAACCUUGAUGAACCAUUAUUAUUUUUGUUUUUCAAAUCAAAUAAAAAU--AAAAACCUACACAC-----AACAAAAGAACUUAUCUCCGCCGGGAUUAUAUCCGCUGGCAG (((.....(((((...)))))((((((((((((........))))))))))--)).......)))..-----...................((((((........).))))).. ( -16.90, z-score = -1.75, R) >droEre2.scaffold_4845 8083618 101 - 22589142 UUCCAUGUUUUUCUUU-UUUAGUACUUAUUUU------UUCCAAUCCAAGUGCAAAUACCUACACAC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAG .....(((........-....((((((.....------.........)))))).............(------(((.((((((....)))))).((((....))))))))))). ( -22.74, z-score = -3.36, R) >droYak2.chr2R 5813920 114 + 21139217 UCAAAUGCUUUUCCUUCUUUAGUAUUUAUUUUUUCCAAUUUAAGUACAAGUACAAAUACAUACACACAGUAAAAGCAAAGGACUUUUGUCCUUCGGGAUUAUUCCCGUUGGCAG .....(((((((.((......(((((((..((....))..)))))))..(((....)))........)).)))))))((((((....))))))(((((....)))))....... ( -26.30, z-score = -4.04, R) >droSec1.super_1 11349838 103 - 14215200 UCAAAUGCUUUUCUUUCUUUAGUAUUUAUUUUC-----GUCCAAUACAAGUACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAG ..(((((((...........)))))))......-----..........................(((------(((.((((((....)))))).((((....)))))))))).. ( -25.40, z-score = -4.21, R) >droSim1.chr2R 12591098 103 - 19596830 UCAAAUGCUUUUCUUUCUUUAGUAUUUAUUUUC-----GUCCAAUACAAGUACAAAUACCUACAGUC------AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAG ..(((((((...........)))))))......-----..........................(((------(((.((((((....)))))).((((....)))))))))).. ( -25.40, z-score = -4.21, R) >consensus UCAAAUGCUUUUCUUUCUUUAGUAUUUAUUUUC_____GUCCAAUACAAGUACAAAUACCUACACAC______AGCAAAGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAG .....((((............((((((....................))))))........................((((((....))))))(((((....)))))..)))). (-13.10 = -13.85 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:31:06 2011