| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,149,232 – 3,149,332 |
| Length | 100 |
| Max. P | 0.918089 |

| Location | 3,149,232 – 3,149,332 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.41235 |
| G+C content | 0.39107 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.19 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
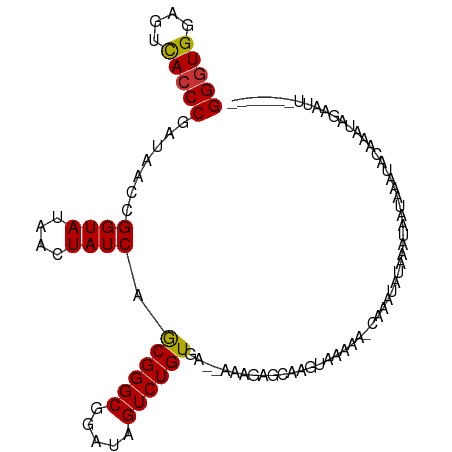
>dm3.chr2L 3149232 100 + 23011544 GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGA--AAAGAGCAAGUAAAAAGCAAAUAUAAAUAAUAAAUACAAAUAGAAUU------- (((((....)))))........((((..((....((((((.....))))))..--..)).((.........))...............))))..........------- ( -18.50, z-score = -1.35, R) >droSim1.chr2L 3091360 100 + 22036055 GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGG--AAAGAGCAAGUAAAAAGCAAAUAUAAAUAAUAAAUACAAAUAGAAUU------- (((((....)))))........((((..((.((.((((((.....)))))).)--).)).((.........))...............))))..........------- ( -20.70, z-score = -1.90, R) >droSec1.super_5 1301671 100 + 5866729 GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGG--AAAGAGCAAGUAAAAAGCAAAUAUAAAUAAUAAAUACAAAUAGAAUU------- (((((....)))))........((((..((.((.((((((.....)))))).)--).)).((.........))...............))))..........------- ( -20.70, z-score = -1.90, R) >droYak2.chr2L 3144903 99 + 22324452 GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGA--AAAGAGCAAGAAAAAG-CUAAUAUAAAUAAUAAAUACAAAUGGAUUU------- (((((....))))).....((.((((..((....((((((.....))))))..--..))(((........)-))..............))))....))....------- ( -21.10, z-score = -1.54, R) >droEre2.scaffold_4929 3193100 99 + 26641161 GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGA--AAAGAGCAAGUAAAAG-CACACAUAAAUAAUAAAUACAAAUAGAAUU------- (((((....)))))........((((..((....((((((.....))))))..--..)).((........)-)...............))))..........------- ( -19.30, z-score = -1.29, R) >dp4.chr4_group4 5125092 82 - 6586962 GGGUGGAGUCACCCGAUAACCGGUAUAGCUAUCAGCGGGCGGAUAGUCUGUAA--AAAGAGUAA-UAAAUA-CAGAAAUUAACUAU----------------------- (((((....))))).....(((.(...((.....)).).)))....((((((.--.........-....))-))))..........----------------------- ( -18.84, z-score = -1.39, R) >droPer1.super_10 2969090 82 - 3432795 GGGUGGAGUCACCCGAUAACCGGUAUAGCUAUCAGCGGGCGGAUAGUCUGUAA--AAAGAGUAA-UAAAUA-AAGAAAUUAACUAU----------------------- (((((....)))))....(((..((((((((((.(....).))))).))))).--...).))..-......-..............----------------------- ( -17.60, z-score = -1.15, R) >droVir3.scaffold_12963 20104591 109 + 20206255 GGGUGGAGUUAACCGAUAACCGGUAUAGCUAUCAACGGGCGGAUAGUCUGUGUGUAUAGAGAGAGAGACCAAUGCAAAUUCGUUAGAUACAUAAUGAAAAUAUACAUAC .(((((.....((((.....))))....))))).((((((.....))))))(((((((....(......)........(((((((......)))))))..))))))).. ( -24.20, z-score = -1.16, R) >consensus GGGUGGAGUCACCCGAUAACCGGUAUAACUAUCAGCGGGCGGAUAGUCUGUGA__AAAGAGCAAGUAAAAA_CAAAUAUAAAUAAUAAAUACAAAUAGAAUU_______ (((((....))))).......((((....)))).((((((.....)))))).......................................................... (-15.28 = -15.19 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:13 2011