| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,825,313 – 13,825,411 |
| Length | 98 |
| Max. P | 0.989498 |

| Location | 13,825,313 – 13,825,411 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Shannon entropy | 0.42529 |
| G+C content | 0.44085 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.01 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 13825313 98 - 21146708 AUUACUUUCACGUCAGUCAUAUGCACUC---GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUCAG---CCGAUAG----GCGCACACACAAAACG------- ...........((.(.((((((((((.(---((....))))))))))))).).))................-......(---((....)----))..............------- ( -27.50, z-score = -3.29, R) >droSim1.chr2R 12561232 98 - 19596830 AUUACUUUCACGUCAGUCAUAUGCACUC---GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUCAG---CCGAUAG----GCGCACACACAAAACG------- ...........((.(.((((((((((.(---((....))))))))))))).).))................-......(---((....)----))..............------- ( -27.50, z-score = -3.29, R) >droSec1.super_1 11319970 98 - 14215200 AUUACUUUCACGUCAGUCAUAUGCACUC---GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUCAU---CCGAUAG----GCGCACACACAAAACG------- ..........(((((.((((((((((.(---((....))))))))))))).)..........(((....))-)......---......)----))).............------- ( -24.40, z-score = -2.63, R) >droYak2.chr2R 5783310 103 + 21139217 AUUACUUUCACGUCAGUCAUAUGCACUC---GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUCAGCCGCCGAUAGG--CGCGCACACACAAAACG------- ...........((.(.((((((((((.(---((....))))))))))))).).))................-......((((((....))--)).))............------- ( -32.80, z-score = -3.97, R) >droEre2.scaffold_4845 8054052 98 - 22589142 AUUACUUUCACGUCAGUCAUAUGCACUC---GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUCAG---CCGAUAG----GCGCACACACAAAACG------- ...........((.(.((((((((((.(---((....))))))))))))).).))................-......(---((....)----))..............------- ( -27.50, z-score = -3.29, R) >droAna3.scaffold_13266 9517703 107 + 19884421 AUUACUUUCACGUCAGUCAUAUGCACUC---GAGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-ACCAUAG---UCGAUAGG--CGCACACCCACACACAGCCAAAUA ...........((.(.((((((((((.(---(......)))))))))))).).))................-.......---......((--(...............)))..... ( -21.66, z-score = -1.42, R) >droMoj3.scaffold_6496 8471324 103 - 26866924 AUUACUUUUACGUCAGUCAUAUGCACCACUCGGGCAGCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUGCCCAUAG---UCAAUGUGCCAGCACAUCAGAACG---------- ....((.(((.((.(.(((((((((((....((....))))))))))))).).)).)))..))................---((.(((((....)))))..))...---------- ( -23.50, z-score = -0.75, R) >droVir3.scaffold_12875 5282191 102 + 20611582 AUUACUUUUACGUCAGUCAUAUGCACCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUGCCCAUAG---CCGAUGCGCCAG-GCACCAGCAUG---------- ....((.(((.((.(.(((((((((((....((....))))))))))))).).)).)))..)).......((((....(---(....))....)-)))........---------- ( -27.10, z-score = -1.23, R) >droGri2.scaffold_15245 8029897 88 - 18325388 AUUACUUUAACGUCAGUCAUAUGCACCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUGCCCAUUG---CCAAUAUGC------------------------- ......((((.((.(.(((((((((((....((....))))))))))))).).))))))....................---.........------------------------- ( -20.30, z-score = -1.28, R) >droWil1.scaffold_180745 2320279 99 - 2843958 AUUACUUUUACGUCACUCAUAUGCACUCGUUGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-ACCAUAG---CCAAUACA---GCACACCCACACA---------- ....((.(((.((.(((((((((((((....((....))))))))))))))).)).)))..))........-......(---(.......---))...........---------- ( -23.30, z-score = -2.72, R) >droPer1.super_2 6501990 92 - 9036312 AUUACUUUUACGUCAGUCAUAUGCAAUA---GGGUACCUUGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUUG----CUGGCAUGAAAAUACAUA---------------- .....(((((.((((((((((((((.((---((....)))))))))))))............(((....))-).....----))))).))))).......---------------- ( -21.10, z-score = -1.77, R) >dp4.chr3 6286280 92 - 19779522 AUUACUUUUACGUCAGUCAUAUGCAAUA---GGGUACCUUGUGCGUAUGAGUAACUUAAUUAGAAAACUUU-CCCUUG----CUGGCAUGAAAAUACAUA---------------- .....(((((.((((((((((((((.((---((....)))))))))))))............(((....))-).....----))))).))))).......---------------- ( -21.10, z-score = -1.77, R) >consensus AUUACUUUCACGUCAGUCAUAUGCACUC___GGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUU_CCCUUAG___CCGAUAGG___GCGCACACACAAA__________ ...........((.(.(((((((((((....((....))))))))))))).).))............................................................. (-16.88 = -17.01 + 0.13)

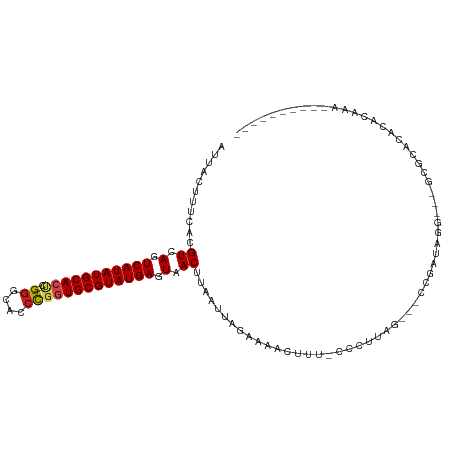
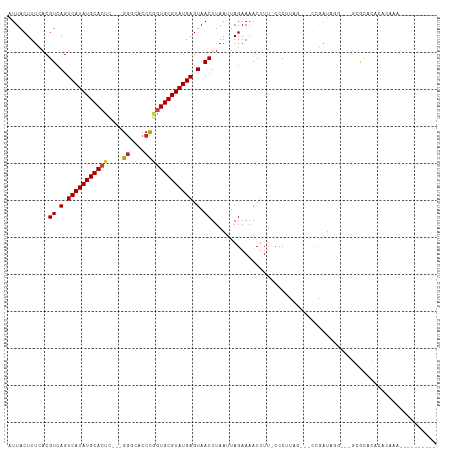
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:59 2011