| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,821,379 – 13,821,497 |
| Length | 118 |
| Max. P | 0.887772 |

| Location | 13,821,379 – 13,821,497 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Shannon entropy | 0.32143 |
| G+C content | 0.42662 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
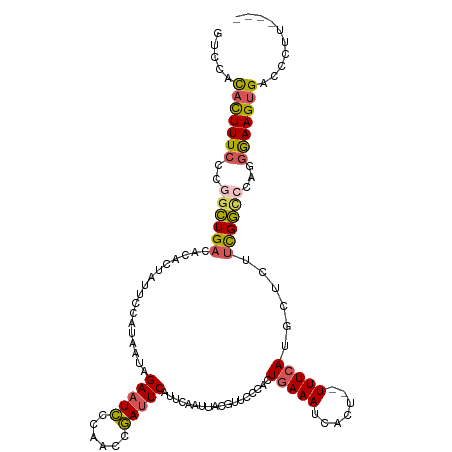
>dm3.chr2R 13821379 118 + 21146708 GUCCACACUUUCCCGGCUGACAGACUAUUCCAUAAUAGAAUCCCAACCGAUUCAUUCAAUUACGUUCCCACUGAAAUCACU--UUUCAUGCUCUUCGGCCCAGGGAAGUGACCCUUUUCC .....((((((((.((((((.(((.............(((((......)))))..................(((((.....--)))))...)))))))))..)))))))).......... ( -28.70, z-score = -3.54, R) >droSim1.chr2R 12557253 114 + 19596830 GUCCACACUUUCCCGGCUGACAGACUAUUCCAUAAUAGAAUCCCAAACGAUUCAUUCAAUUACGUUCCCACUGAAAUCACU--UUUCAUGCUCUUCGGUCCAUGGAAGUGACCCUU---- .....(((((((..((((((.(((.............(((((......)))))..................(((((.....--)))))...)))))))))...)))))))......---- ( -22.60, z-score = -1.76, R) >droSec1.super_1 11316136 114 + 14215200 GUCCACACUUUCCCGGCUGACACACUAUUCCAUAAUAGAAUCCCAAACGAUUCAUUCAAUUACGUUCCCACUGAAAUCACU--UUUCAUGCUCUUCGGUCCAUGGAAGUGACCCUU---- .....(((((((..((((((.................(((((......)))))..................(((((.....--)))))......))))))...)))))))......---- ( -21.10, z-score = -1.77, R) >droEre2.scaffold_4845 8050057 118 + 22589142 GUCCACACUUUCCCGGCUGACAAACUAUUCCAUAAUAGAAUCCCAACCGAUUCAUUCAAUUACUUUCCCACUGAAAUCACU--UUUCAUGCUCUUCGGCACAGGGAAGUGACCCUUUUUU .....((((((((.((((((.................(((((......)))))..................(((((.....--)))))......))))).).)))))))).......... ( -25.20, z-score = -3.30, R) >droAna3.scaffold_13266 9513422 112 - 19884421 -CCCUUAUUUUGACGCUUGUCCCACUAUUUAAUAAUAGAUUUCUAACCAAUUCAUUCACUAAC---CCCACUCAAAUCACACAUUUUAUGCUCUUUGACGUGGAAAAGGGUUCGAU---- -(((((.....(((....)))...(((((....))))).........................---.(((((((((...((.......))...))))).))))..)))))......---- ( -16.00, z-score = -0.84, R) >consensus GUCCACACUUUCCCGGCUGACACACUAUUCCAUAAUAGAAUCCCAACCGAUUCAUUCAAUUACGUUCCCACUGAAAUCACU__UUUCAUGCUCUUCGGCCCAGGGAAGUGACCCUU____ .....(((((((..((((((.................(((((......)))))..................(((((.......)))))......))))))...))))))).......... (-14.00 = -14.16 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:58 2011