| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,809,115 – 13,809,194 |
| Length | 79 |
| Max. P | 0.745620 |

| Location | 13,809,115 – 13,809,194 |
|---|---|
| Length | 79 |
| Sequences | 9 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 67.25 |
| Shannon entropy | 0.63744 |
| G+C content | 0.37696 |
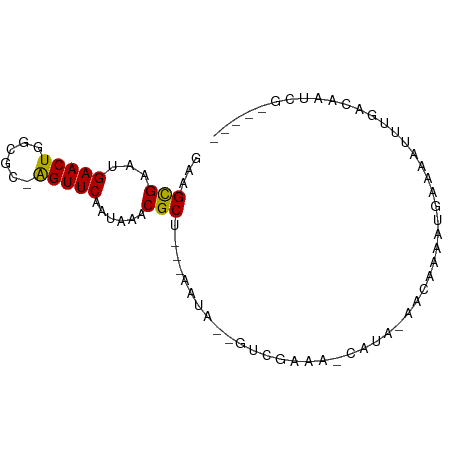
| Mean single sequence MFE | -12.90 |
| Consensus MFE | -5.87 |
| Energy contribution | -5.60 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
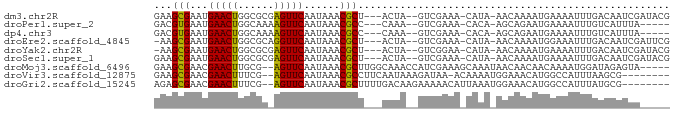
>dm3.chr2R 13809115 79 - 21146708 GAAGCGAAUGAACUGGCGCGAGUUCAAUAAACGCU---ACUA--GUCGAAA-CAUA-AACAAAAUGAAAAUUUGACAAUCGAUACG ((((((..((((((......)))))).....))))---....--((((((.-(((.-......)))....))))))..))...... ( -13.00, z-score = -1.13, R) >droPer1.super_2 6482368 74 - 9036312 GACGUGAAUGAACUGGCAAAAGUUCAAUAAACGCC---CAAA--GUCGAAA-CACA-AGCAGAAUGAAAAUUUGUCAUUUA----- ((((((..((((((......)))))).....(((.---....--).))...-))).-....((((....))))))).....----- ( -13.10, z-score = -1.11, R) >dp4.chr3 6267708 74 - 19779522 GACGUGAAUGAACUGGCAAAAGUUCAAUAAACGCC---CAAA--GUCGAAA-CACA-AGCAGAAUGAAAAUUUGUCAUUUA----- ((((((..((((((......)))))).....(((.---....--).))...-))).-....((((....))))))).....----- ( -13.10, z-score = -1.11, R) >droEre2.scaffold_4845 8037809 78 - 22589142 -AAGCGAAUGAACUGGCGCAGGUUCAAUAAACGCU---ACUA--GUCGAAA-CAUA-AACAAAAUGGAAAUUUGACAAUCGAUUCG -.((((..((((((......)))))).....))))---....--((((((.-(((.-......)))....)))))).......... ( -14.10, z-score = -1.19, R) >droYak2.chr2R 5766367 78 + 21139217 -AAGCGAAUGAACUGGCGCGAGUUCAAUAAACGCU---ACUA--GUCGGAA-CAUA-AACAAAAUGAAAAUUUGACAAUCGAUACG -.((((..((((((......)))))).....))))---....--((((((.-(((.-......)))....)))))).......... ( -12.50, z-score = -0.69, R) >droSec1.super_1 11304076 79 - 14215200 GAAGCGAAUGAACUGGCGCGAGUUCAAUAAACGCU---ACUA--GUCGAAA-CAUA-AACAAAAUGAAAAUUUGACAAUCGAUACG ((((((..((((((......)))))).....))))---....--((((((.-(((.-......)))....))))))..))...... ( -13.00, z-score = -1.13, R) >droMoj3.scaffold_6496 8445024 79 - 26866924 GAAGCGAACGAACUUGCG--AGUUCAAUAAACGCUUGGCAAACCAUCGAAAGCAAAUAACAACAACAAAAUGGAUAGAGUA----- ...((....(((((....--)))))......((..(((....))).))...))............................----- ( -9.90, z-score = 0.13, R) >droVir3.scaffold_12875 5255129 75 + 20611582 GAAGCGAACGAACUUUCG--AGUUCAAUAAACGCCUUCAAUAAAGAUAA-ACAAAAUGGAAACAUGGCCAUUUAAGCG-------- ...((....(((((....--))))).......(((.((......))...-.....(((....)))))).......)).-------- ( -13.80, z-score = -2.08, R) >droGri2.scaffold_15245 6166769 76 + 18325388 AGAGCGAACGAACUUUCG--AGUUCAAUAAACGCUUUUGACAAGAAAAACAUUAAAUGGAAACAUGGCCAUUUAUGCG-------- ((((((...(((((....--)))))......))))))...............(((((((........)))))))....-------- ( -13.60, z-score = -1.31, R) >consensus GAAGCGAAUGAACUGGCGC_AGUUCAAUAAACGCU___AAUA__GUCGAAA_CAUA_AACAAAAUGAAAAUUUGACAAUCG_____ ...(((...(((((......)))))......))).................................................... ( -5.87 = -5.60 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:56 2011