
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 13,802,696 – 13,802,798 |
| Length | 102 |
| Max. P | 0.761112 |

| Location | 13,802,696 – 13,802,798 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.60600 |
| G+C content | 0.56044 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -15.17 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
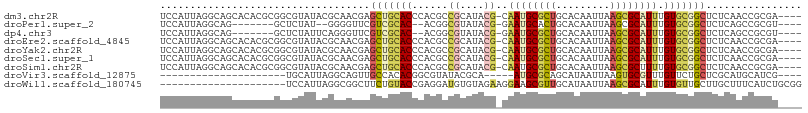
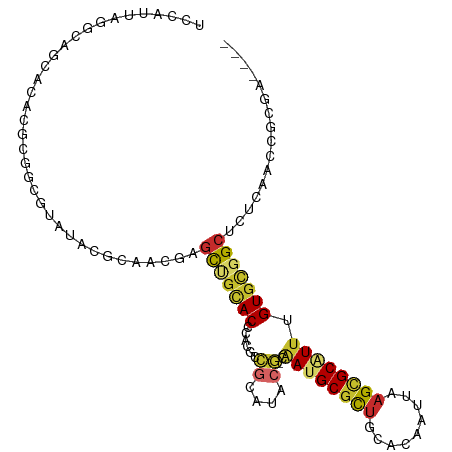
>dm3.chr2R 13802696 102 + 21146708 UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG-CAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAACCGCGA---- ........(((((...((..(((....)))..)).)))))......((((((((..-.((((((((.........))))))))))))))))............---- ( -39.90, z-score = -2.05, R) >droPer1.super_2 6476435 91 + 9036312 UCCAUUAGGCAG-------GCUCUAU--GGGGUUCGUCGCAC--ACGGCGUAUACG-GAAUGCACUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAGCCGCGU---- (((((..((...-------.))..))--)))((.(((((...--.)))))...)).-((((((.((.........)).))))))..(((((.....)))))..---- ( -29.10, z-score = -0.28, R) >dp4.chr3 6261750 93 + 19779522 UCCAUUAGGCAG-------GCUCUAUUCAGGGUUCGUCGCAC--ACGGCGUAUACG-GAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAGCCGCGU---- .......(((.(-------(((((....)))))).(((((((--.((.......))-(((((((((.........)))))))))))))))).....)))....---- ( -34.90, z-score = -2.06, R) >droEre2.scaffold_4845 8031426 102 + 22589142 UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG-CAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAACCGCGA---- ........(((((...((..(((....)))..)).)))))......((((((((..-.((((((((.........))))))))))))))))............---- ( -39.90, z-score = -2.05, R) >droYak2.chr2R 5759827 102 - 21139217 UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG-CAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAACCGCGA---- ........(((((...((..(((....)))..)).)))))......((((((((..-.((((((((.........))))))))))))))))............---- ( -39.90, z-score = -2.05, R) >droSec1.super_1 11297785 102 + 14215200 UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG-CAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAACCGCGA---- ........(((((...((..(((....)))..)).)))))......((((((((..-.((((((((.........))))))))))))))))............---- ( -39.90, z-score = -2.05, R) >droSim1.chr2R 12545428 102 + 19596830 UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG-CAAUGCGCUGCACAAUUAAGCGCUUUUGUGCGGCUCUCAACCGCGA---- ........(((((...((..(((....)))..)).))))).....(((.((((...-..))))(((((((((..........)))))))))........))).---- ( -36.30, z-score = -0.89, R) >droVir3.scaffold_12875 5247971 77 - 20611582 ---------------------UGCAUUAGGCAGUUGCCACACGGCGUAUACGCA-----AUGCGCAGCAUAAUUAAGUGCGUUUGUUCUGCUCGCAUGCAUCG---- ---------------------(((((..(((((....((.((.((((((.....-----)))))).((((......)))))).))..)))))...)))))...---- ( -23.40, z-score = 0.27, R) >droWil1.scaffold_180745 2290342 86 + 2843958 ---------------------UCCAUUAGGCGGCUUCUGUACCGAGGAUGUGUAGAAGGAAGCGUUGCAUAAUUAAGCGCAUUUGUGUUGCUUGCUUUCAUCUGCGG ---------------------..((((...(((........)))..))))((((((.(((((((..(((.......((((....))))))).))))))).)))))). ( -25.71, z-score = -0.77, R) >consensus UCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACG_CAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCUCUCAACCGCGA____ .....................................(((.........((((......))))(((((((((..........)))))))))........)))..... (-15.17 = -14.76 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:30:55 2011