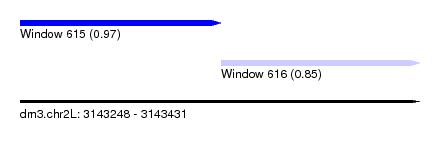
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,143,248 – 3,143,431 |
| Length | 183 |
| Max. P | 0.965930 |

| Location | 3,143,248 – 3,143,340 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 54.64 |
| Shannon entropy | 0.64021 |
| G+C content | 0.50034 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -9.98 |
| Energy contribution | -9.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
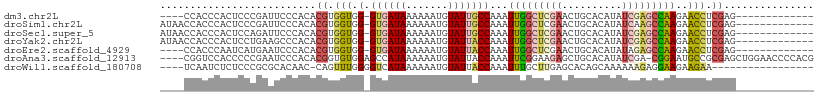
>dm3.chr2L 3143248 92 + 23011544 GAGCAAAACAAACCUGU-CACGAGCGAAAGUUCUUCAAGCAGACACGCGACAGUGGUUGGAUGGGCACACCACCCAGAGUCACCCAUAACCAU- .............((((-(..((((....)))).....((......)))))))(((.(((.((((.......))))...))).))).......- ( -25.30, z-score = -2.07, R) >droEre2.scaffold_4929 3187122 84 + 26641161 GGGCAAAACAAACCUGU-CACUAGCGAAAGUUCUUCAGUUAGACACGCGACAGUGGUUGGGUGGGCUCACCACCCACAGCCAGCC--------- .(((.........((((-(....((....))(((......))).....)))))(((((((((((.....)))))))..)))))))--------- ( -30.90, z-score = -2.44, R) >droWil1.scaffold_180708 1068707 81 + 12563649 -----------GUCAAUGCAGCAACUGAAAUACAUUGAACAGACACGCGAC--UGGCUGACAUAGAAAAUUUCCUUUACUCGGCCAAUACAUUC -----------.(((((((((...))).....)))))).............--(((((((...((........))....)))))))........ ( -13.30, z-score = -0.92, R) >consensus G_GCAAAACAAACCUGU_CACCAGCGAAAGUUCUUCAAACAGACACGCGACAGUGGUUGGAUGGGCACACCACCCACAGUCAGCCA__AC__U_ ......................................................((((((.((((.......))))...))))))......... ( -9.98 = -9.77 + -0.22)


| Location | 3,143,340 – 3,143,431 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Shannon entropy | 0.46090 |
| G+C content | 0.47535 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3143340 91 + 23011544 ----CCACCCACUCCCGAUUCCCACACGUGGUGG-GUGAUAAAAAAUGUAUUGCCAAAUUGGCUCGAACUGCACAUAUCGAGCCAAGAACCUCGAG------------- ----.((((((((..((.........)).)))))-)))....................(((((((((..........)))))))))..........------------- ( -26.20, z-score = -2.86, R) >droSim1.chr2L 3084451 95 + 22036055 AUAACCACCCACUCCCGAUUCCCACACGUGGUGG-GUGAUAAAAAAUGUAUUGCCAAAUUGGCUCGAACUGCACAUAUCAAGCCAAGAACCUCGAG------------- .....((((((((..((.........)).)))))-)))....................((((((.((..........)).))))))..........------------- ( -20.30, z-score = -1.00, R) >droSec1.super_5 1294999 95 + 5866729 AUAACCACCCACUCCAGAUUCCCACACGUGGUGG-GUGAUAAAAAAUGUAUUGCCAAAUUGGCUCGAACUGCACAUAUCGAGCCAAGAACCUCGAG------------- .....(((((((..(............)..))))-)))....................(((((((((..........)))))))))..........------------- ( -26.40, z-score = -2.85, R) >droYak2.chr2L 3136143 95 + 22324452 AUAACCACCCACUCCUGAAGCCCACACGUGGUGG-GUGAUAAAAAAUGUAUUACCAAAUUGGCUCGAACUGCACAUAUCGAGCCAAGAACCUCGAG------------- .....(((((((..(((.......)).)..))))-)))....................(((((((((..........)))))))))..........------------- ( -27.20, z-score = -3.43, R) >droEre2.scaffold_4929 3187206 91 + 26641161 ----CCACCCAAUCAUGAAUCCCACACGUGGUGG-GUGAUAAAAAAUGUAUUACCAAAUUGGCUCGAACUGCACAUAUAGAGCCAAGAACCUCGAG------------- ----.((((((.(((((.........))))))))-)))....................(((((((..............)))))))..........------------- ( -23.24, z-score = -2.46, R) >droAna3.scaffold_12913 363685 104 + 441482 ----CGGUCCACCCCCGAAUCCCACACGGUGUGGAGCCAUAAAAAAUGUAUUACCAAAUUCGGAAGAGCUGCACAUAUCGA-CGGAAUGCCGCGAGCUGGAACCCCACG ----.((((((.................(((((.((((((.....)))...........((....))))).))))).(((.-(((....))))))..))).)))..... ( -25.30, z-score = -0.22, R) >droWil1.scaffold_180708 1068788 87 + 12563649 ----UCAAUCUCUCCCGCGCACAAC-CAGUUUGGGGUCAUAAAAAAUGUAUUACCAAAUUUGCUUGAGCACAGCAAAAAAGAGGAAGAAGAA----------------- ----....(((((.(((.((.((((-.(((((((...(((.....))).....))))))).).))).)).)..(......).)).)).))).----------------- ( -14.20, z-score = 0.36, R) >consensus ____CCACCCACUCCCGAAUCCCACACGUGGUGG_GUGAUAAAAAAUGUAUUACCAAAUUGGCUCGAACUGCACAUAUCGAGCCAAGAACCUCGAG_____________ ..........................((.(((.(.((((((.......)))))))...(((((((((..........)))))))))..))).))............... (-12.29 = -12.87 + 0.58)
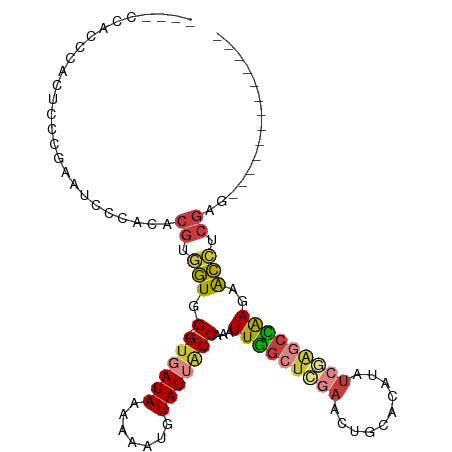
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:12 2011